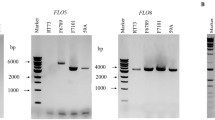
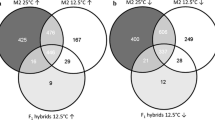
Abstract
Most commercial yeast strains are nonflocculent. However, controlled flocculation phenotypes could provide significant benefits to many fermentation-based industries. In nonflocculent laboratory strains, it has been demonstrated that it is possible to adjust flocculation and adhesion phenotypes to desired specifications by altering expression of the otherwise silent but dominant flocculation (FLO) genes. However, FLO genes are characterized by high allele heterogeneity and are subjected to epigenetic regulation. Extrapolation of data obtained in laboratory strains to industrial strains may therefore not always be applicable. Here, we assess the adhesion phenotypes that are associated with the expression of a chromosomal copy of the FLO1, FLO5, or FLO11 open reading frame in two nonflocculent commercial wine yeast strains, BM45 and VIN13. The chromosomal promoters of these genes were replaced with stationary phase-inducible promoters of the HSP30 and ADH2 genes. Under standard laboratory and wine making conditions, the strategy resulted in expected and stable expression patterns of these genes in both strains. However, the specific impact of the expression of individual FLO genes showed significant differences between the two wine strains and with corresponding phenotypes in laboratory strains. The data suggest that optimization of the flocculation pattern of individual commercial strains will have to be based on a strain-by-strain approach.









Similar content being viewed by others
References
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1995) Current protocols in molecular biology. Wiley, New York
Bely M, Sablayrolles JM, Barre P (1990) Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in enological conditions. J Ferment Bioeng 70:246–252
Bony M, Thines-Sempoux D, Barre P, Blondin B (1997) Localization and cell surface anchoring of the Saccharomyces cerevisiae flocculation protein Flo1p. J Bacteriol 179:4929–4936
Caro LH, Tettelin H, Vossen JH, Ram AF, van den Ende H, Klis FM (1997) In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477–1489
Chambers P, Issaka A, Palecek SP (2004) Saccharomyces cerevisiae JEN1 promoter activity is inversely related to concentration of repressing sugar. Appl Environ Microbiol 70:8–17
Ciriacy M (1997) Alcohol dehydrogenases. In: Zimmermann FK, Entian K-D (eds) Yeast sugar metabolism: biochemistry, genetics, biotechnology and applications. Technomic, Lancaster, pp 213–223
Cunha AF, Missawa SK, Gomes LH, Reis SF, Pereira GA (2006) Control by sugar of Saccharomyces cerevisiae flocculation for industrial ethanol production. FEMS Yeast Res 6:280–287
D’Hautcourt O, Smart KA (1999) The measurement of brewing yeast flocculation. J Am Soc Brew Chem 57:123–128
De Groot PW, Hellingwerf KJ, Klis FM (2003) Genome-wide identification of fungal GPI proteins. Yeast 20:781–796
Du Preez JC, Mare JE, Albertyn J, Kilian SG (2001) Transcriptional repression of ADH2-regulated beta-xylanase production by ethanol in recombinant strains of Saccharomyces cerevisiae. FEMS Yeast Res 1:233–240
Fichtner L, Schulze F, Braus GH (2007) Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell–cell and cell-substrate adherence of S. cerevisiae S288C. Mol Microbiol 66:1276–1289
Fidalgo M, Barrales RR, Ibeas JI, Jimenez J (2006) Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci USA 103:11228–11233
Govender P, Domingo JL, Bester MC, Pretorius IS, Bauer FF (2008) Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae. Appl Environ Microbiol 74:6041–6052
Guo B, Styles CA, Feng Q, Fink GR (2000) A Saccharomyces gene family involved in invasive growth, cell–cell adhesion, and mating. Proc Natl Acad Sci USA 97:12158–12163
Hamada K, Fukuchi S, Arisawa M, Baba M, Kitada K (1998) Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol Gen Genet 258:53–59
Hinchcliffe E, Box WG, Walton EF, Appleby M (1985) The influences of cell wall hydrophobicity on the top fermenting properties of brewing yeast. Proc Eur Brew Congr 20:323–330
Ishigami M, Nakagawa Y, Hayakawa M, Iimura Y (2004) FLO11 is essential for flor formation caused by the C-terminal deletion of NRG1 in Saccharomyces cerevisiae. FEMS Microbiol Lett 237:425–430
James TC, Campbell SG, Bond UM (2002) Comparative analysis of global gene expression in lager and laboratory yeast strains grown in wort. Proc IEEE 90:1887–1899
Kealey JT, Liu L, Santi DV, Betlach MC, Barr PJ (1998) Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc Natl Acad Sci USA 95:505–509
Kjeldsen T (2000) Yeast secretory expression of insulin precursors. Appl Microbiol Biotechnol 54:277–286
Kobayashi O, Hayashi N, Kuroki R, Sone H (1998) Region of FLO1 proteins responsible for sugar recognition. J Bacteriol 180:6503–6510
Lee KM, DaSilva NA (2005) Evaluation of the Saccharomyces cerevisiae ADH2 promoter for protein synthesis. Yeast 22:431–440
Lipke PN, Ovalle R (1998) Cell wall architecture in yeast: new structure and new challenges. J Bacteriol 180:3735–3740
Manthey GM, Navarro MS, Bailis AM (2004) DNA fragment transplacement in Saccharomyces cerevisiae: some genetic considerations. Methods Mol Biol 262:157–172
Maury J, Asadollahi MA, Moller K, Clark A, Nielsen J (2005) Microbial isoprenoid production: an example of green chemistry through metabolic engineering. Adv Biochem Eng Biotechnol 100:19–51
Ness F, Lavallée F, Dubourdieu D, Aigle M, Dulau L (1993) Identification of yeast strains using the polymerase chain reaction. J Sci Food Agric 62:89–94
Nevoigt E, Kohnke J, Fischer CR, Alper H, Stahl U, Stephanopoulos G (2006) Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl Environ Microbiol 72:5266–5273
Noronha SB, Kaslow DC, Shiloach J (1998) Transition phase in the production of recombinant proteins in yeast under the ADH2 promoter: an important step for reproducible manufacturing of a malaria transmission blocking vaccine candidate. J Ind Microbiol 20:192–199
Pretorius IS (2000) Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675–729
Pretorius IS, Bauer FF (2002) Meeting the consumer challenge through genetically customized wine-yeast strains. Trends Biotechnol 20:426–432
Rossouw D, Naes T, Bauer FF (2008) Linking gene regulation and the exo-metabolome: a comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genomics 9:530
Saitoh S, Ishida N, Onishi T, Tokuhiro K, Nagamori E, Kitamoto K, Takahashi H (2005) Genetically engineered wine yeast produces a high concentration of L-lactic acid of extremely high optical purity. Appl Environ Microbiol 71:2789–2792
Sambrook KJ, Fritsch EF, Maniatis T (1989) Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor
Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latgé JP, Fink GR, Foster KR, Verstrepen KJ (2008) FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135:726–737
Stratford M (1989) Evidence for two mechanisms of flocculation in Saccharomyces cerevisiae. Yeast 5:S441–S445
Stratford M, Assinder S (1991) Yeast flocculation: Flo1 and NewFlo phenotypes and receptor structure. Yeast 7:559–574
Teunissen AW, Steensma HY (1995) Review: the dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 11:1001–1013
Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA (2005) Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet 37:630–635
Van Mulders SE, Christianen E, Saerens SM, Daenen L, Verbelen PJ, Willaert R, Verstrepen KJ, Delvaux FR (2009) Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res 9:178–190
Varela C, Cardenas J, Melo F, Agosin E (2005) Quantitative analysis of wine yeast gene expression profiles under winemaking conditions. Yeast 22:369–383
Verstrepen KJ, Klis FM (2006) Flocculation, adhesion and biofilm formation in yeasts. Mol Microbiol 60:5–15
Verstrepen KJ, Michiels C, Derdelinckx G, Delvaux FR, Winderickx J, Thevelein JM, Bauer FF, Pretorius IS (2001) Late fermentation expression of FLO1 in Saccharomyces cerevisiae. J Am Soc Brew Chem 59:69–76
Verstrepen KJ, Reynolds TB, Fink GR (2004) Origins of variation in the fungal cell surface. Nat Rev Microbiol 2:533–540
Verstrepen KJ, Jansen A, Lewitter F, Fink GR (2005) Intragenic tandem repeats generate functional variability. Nat Genet 37:986–990
Wang D, Wang Z, Liu N, He X, Zhang B (2008) Genetic modification of industrial yeast strains to obtain controllable NewFlo flocculation property and lower diacetyl production. Biotechnol Lett 30:2013–2018
Watari J, Takata Y, Ogawa M, Murakami J, Koshino S (1991) Breeding of flocculent industrial Saccharomyces cerevisiae strains by introducing the flocculation gene FLO1. Agric Biol Chem 55:1547–1552
Wills C (1976) Production of yeast alcohol dehydrogenase isoenzymes by selection. Nature 261:26–29
Winston F, Dollard C, Ricupero-Hovasse SL (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53–55
Acknowledgments
This work was financially supported by the National Research Foundation (NRF) and the South African Wine Industry (Winetech).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Govender, P., Bester, M. & Bauer, F.F. FLO gene-dependent phenotypes in industrial wine yeast strains. Appl Microbiol Biotechnol 86, 931–945 (2010). https://doi.org/10.1007/s00253-009-2381-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2381-1