Abstract
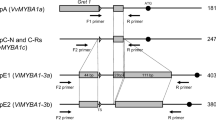
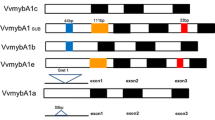
The genetics and biochemistry of anthocyanins and flavonol biosynthesis and their role in plant organ pigmentation is well established in model species. However, the genetic basis of colour variation is species specific and understanding this variation is very relevant in many fruit and flower crop species. Among grape cultivars, there is a wide genetic variation for berry colour ranging from yellow-green (“white” cultivars) to dark blue berries. Berry colour results from the synthesis and accumulation of anthocyanins in the berry skin, which in plants is commonly regulated by transcription factors belonging to the MYB and bHLH families. In this work, we aimed to identify the major genetic determinants of berry colour variation in a large collection of table grape cultivars and somatic variants. The genetic analyses of berry colour in a few grape segregating progenies had previously identified a single locus on linkage group 2 responsible for colour variation. Furthermore, somatic variation for berry skin colour in cultivar Italia had been associated with the presence of a Gret1 retrotransposon in the promoter region of VvmybA1, a Myb gene whose expression is associated to skin colouration. The results show that VvmybA1 is the gene underlying the mapped locus controlling berry colour in grape. Additionally, the molecular analyses indicate that genetic and somatic berry colour variation can be associated to molecular variation at VvmybA1 in more than 95% of the analyzed cultivars. Thus, VvmybA1 is a major determinant of berry colour variation in table grape and its instability is the major cause of somatic variation for this trait.





Similar content being viewed by others
References
Adam-Blondon AF, Roux C, Claux D, Butterlin G, Merdinoglu D, This P (2004) Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor Appl Genet 109:1017–1027
Arakawa O, Shinoda M, Hiraga M, Wang H (1999) Comparison of anthocyanin synthesis of true-to-type ‘Tsugaru’ apple and its red sport strains. J Hortic Sci Biotechnol 74:738–742
Awad MA, de Jager A, van Westing LM (2000) Flavonoid and chlorogenic acid levels in apple fruit: characterisation of variation. Sci Hortic-Amsterdam 83:249–263
Ben-Yehudah G, Korchinsky R, Redel G, Ovadya R, Oren-Shamir M, Cohen Y (2005) Colour accumulation patterns and the anthocyanin biosynthetic pathway in ‘Red Delicious’ apple variants. J Hortic Sci Biotechnol 80:187–192
Bogs J, Ebadi A, McDavid D, Robinson SP (2006) Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol 140:279–291
Borovsky Y, Oren-Shamir M, Ovadia R, De Jong W, Paran I (2004) The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to anthocyanin2 of petunia. Theor Appl Genet 109:23–29
Boss P, Davies C, Robinson S (1996a) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111:1059–1066
Boss P, Davies C, Robinson S (1996b) Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol Biol 32:565 - 569
Cabezas JA, Cervera MT, Arroyo-Garcia R, Ibanez J, Rodriguez-Torres I, Borrego J, Cabello F, Martinez-Zapater JM (2003) Garnacha and Garnacha Tintorera: genetic relationships and the origin of teinturier varieties cultivated in Spain. Am J Enol Vitic 54:237–245
Carreño J, Martinez A, Almela L, Fernandez-Lopez JA (1995) Proposal of an index for the objective evaluation of the colour of red table grapes. Food Res Inter 28:373–377
Castellarin SD, Di Gaspero G, Marconi R, Nonis A, Peterlunger E, Paillard S, Adam-Blondon AF, Testolin R (2006) Colour variation in red grapevines (Vitis vinifera L.): genomic organisation, expression of flavonoid 3′-hydroxylase, flavonoid 3′, 5′-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin. BMC Genomics 7:12
Doligez A, Bouquet A, Danglot Y, Lahogue F, Riaz S, Meredith CP, Edwards KJ, This P (2002) Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor Appl Genet 105:780–795
van Dyk MM, Koning G, Simayi Z, Booi S, Maharaj R, Selala MC, du Preez MG, Rees DJG, Labuschagné IF, Warnich L (2005) Molecular genetic studies on pears (Pyrus spp.) in the Western Cape. Acta Hortic 671:259–265
Fisher BM, Salakhutdinov I, Akkur M, Eibach R, Edwards KJ, Töpfer R, Zyprian EM (2004) Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor Appl Genet 108:501–515
Franks T, Botta R, Thomas MR, Franks J (2002) Chimerism in grapevines: implications for cultivar identity, ancestry and genetic improvement. Theor Appl Genet 104:192–199
Gautier-Hion A, Duplantier J-M, Quris R, Feer F, Sourd C, Decoux J-P, Dubost G, Emmons L, Erard C, Hecketsweiler P, Moungazi A, Roussilhon C, Thiollay J-M (1985) Fruit characters as a basis of fruit choice and seed dispersal in a tropical forest vertebrate community. Oecologia (Historical Archive) 65:324–337
Goff SA, Cone KC, Fromm ME (1991) Identification of functional domains in the maize transcriptional activator C1: comparison of wild-type and dominant inhibitor proteins. Genes Dev 2:298–309
Hall T (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hocquigny S, Pelsy F, Dumas V, Kindt S, Heloir MC, Merdinoglu D (2004) Diversification within grapevine cultivars goes through chimeric states. Genome 47:579–589
Holton T, Cornish E (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083
Jaakola L, Määttä K, Pirttilä A, Törrönen R, Kärenlampi S, Hohtola A (2002) Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol 130:729–739
James WC, Howard BF (1989) The citrus industry. Volumen 2: genetics, breeding, and nucellar embryony. Chapter 5: genetics, breeding, and nucellar embryony. University of California board of regents, Davis
Kevan PG, Baker HG (1983) Insects as flower visitors and pollinators. Annual Review of Entomology 28:407–453
Kobayashi S, Ishimaru M, Ding C, Yakushiji H, Goto N (2001) Comparison of UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci 160:543–550
Kobayashi S, Ishimaru M, Hiraoka K, Honda C (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215:924–933
Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin colour. Science 304:982–982
Kobayashi S, Yamamoto NG, Hirochika H (2005) Association of VvmybA1 gene expression with anthocyanin production in grape (Vitis vinifera) skin-color mutants. J Jpn Soc Hort Sci 74:196–203
Ma JX, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
Ozeki Y, Chikagawa Y, Kimura S, Soh H, Maeda K, Pornsiriwong W, Kato M, Akimoto H, Oyanagi M, Fukuda T, Koda T, Itoh Y, Yamada A, Davies E, Ueno H, Takeda J (2003) Putative cis-elements in the promoter region of the carrot phenylalanine ammonia-lyase gene induced during anthocyanin synthesis. J Plant Res 116:155–159
Quattrocchio F, Wing J, Leppen H, Mol J, Koes R (1993) Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5:1497–1512
Ramsay N, Walker A, Mooney M, Gray J (2003) Two basic-helix-loop-helix genes (MYC-146 and GL3) from Arabidopsis can activate anthocyanin biosynthesis in a white-flowered Matthiola incana mutant. Plant Mol Biol 52:679–688
Robbins M, Paolocci F, Hughes J, Turchetti V, Allison G, Arcioni S, Morris P, Damiani F (2003) Sn, a maize bHLH gene, modulates anthocyanin and condensed tannin pathways in Lotus corniculatus. J Exp Bot 54:239–248
Sainz M, Grotewold E, Chandler V (1997) Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related MYB domain proteins. Plant Cell 9:611–625
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18:831–851
Spelt C, Quattrocchio F, Mol J, Koes R (2000) anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12:1619–1632
This P, Jung A, Boccacci P, Borrego J, Botta R, Costantini L, Crespan M, Dangl GS, Eisenheld C, Ferreira-Monteiro F, Grando S, Ibañez J, Lacombe T, Laucou V, Magalhaes R, Meredith CP, Milani N, Peterlunger E, Regner F, Zulini L, Maul E (2004) Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor Appl Genet 109:1448–1458
Thompson MM, Olmo HP (1963) Cytohistological studies of cytochimeric and tetraploid grapes. Am J Bot 50:901–906
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colourful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493
Zeng Y, Yang T (2002) RNA isolation from highly viscous samples rich in polyphenols and polysaccharides. Plant Mol Biol Rep 20:417a–417e
Acknowledgment
We thank Juan Negueroles (AFREXPORT S.A.) and María del Carmen Martínez (Misión Biológica de Galicia-CSIC) for some of the plant materials used in DNA and RNA analyses. We also thank Cheo Machín for careful editing of the manuscript. This work was funded by a grant from the CDTI (Spanish Ministry of Science and Technology).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Hagemann
Nucleotide sequence data reported are available in the GenBank databases under the accession numbers: DQ403722, DQ345541, DQ345539 and DQ403721.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lijavetzky, D., Ruiz-García, L., Cabezas, J.A. et al. Molecular genetics of berry colour variation in table grape. Mol Genet Genomics 276, 427–435 (2006). https://doi.org/10.1007/s00438-006-0149-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-006-0149-1