-
PDF
- Split View
-
Views
-
Cite
Cite
Antonio J. Castro, Christine Carapito, Nathalie Zorn, Christian Magné, Emmanuelle Leize, Alain Van Dorsselaer, Christophe Clément, Proteomic analysis of grapevine (Vitis vinifera L.) tissues subjected to herbicide stress, Journal of Experimental Botany, Volume 56, Issue 421, November 2005, Pages 2783–2795, https://doi.org/10.1093/jxb/eri271
Close - Share Icon Share
Abstract
Two-dimensional gel electrophoresis coupled to mass spectrometry analysis was used to examine for the first time the effect of a herbicide (flumioxazin) on a crop species (Vitis vinifera L.) at the proteome level. Examination of 2-D maps derived from chemically stressed tissues revealed the presence of 33 spots displaying a differential expression pattern. The presence of stress responsive proteins in the different plant organs analysed suggests that flumioxazin could act systemically. Among the responsive proteins, some photosynthesis-related proteins, including several fragments of the enzyme Rubisco, were identified. This effect suggests that photosynthesis could be impaired by the herbicide. The induction of several enzymatic antioxidant systems was also observed, probably as a result of an oxidative stress. Moreover, the photorespiration pathway was stimulated, as suggested by the induction of some key enzymes involved in this process. Changes in carbon metabolism-associated proteins presumably reflect altered patterns of carbon flux in response to impaired photosynthesis and an increased need for osmotic adjustment in affected tissues. Finally, plant defences were stimulated as revealed by the induction of a set of proteins belonging to the pathogenesis-related 10 class, suggesting that they could play an essential role in cell defence mechanisms against flumioxazin.
Introduction
The ability of plants to cope with a variety of chemical and physical stressors depends on a number of proteins, which are up- and down-regulated as a result of altered gene expression. Most of these molecules display an essential function either in the regulation of the response (e.g. components of the signal transduction pathway) or in the adaptation process (e.g. enzymes involved in stress repair and degradation of damaged cellular contents), allowing plants to recover and survive the stress. Many of these proteins are constitutively expressed under normal conditions but, under a period of stress, they undergo a modification of their expression levels. Besides, some others are specifically synthesized or repressed depending on the stress. Therefore, they may be useful in stress monitoring, providing valuable information about the nature of the stress factor, as well as the physiological state of a biological system. In recent years, proteomic-based technologies have been successfully applied for the systematic scrutiny of the induced gene products in a number of plant species subjected to a wide range of abiotic challenges, including drought (Costa et al., 1998; Riccardi et al., 1998; Salekdeh et al., 2002a, b), anoxia (Chang et al., 2000), salt stress (Salekdeh et al., 2002b), low and high temperature (Lund et al., 1998; Ukaji et al., 1999), starvation (Suzuki et al., 1998), gaseous air pollutants (Agrawal et al., 2002; Rakwal et al., 2003), UV radiation (Rakwal et al., 1999), and heavy metals (Hajduch et al., 2001; Fecht-Christoffers et al., 2003).
Although herbicides have largely contributed to the improvement of crop productivity in terms of yield and quality, their improper use is currently becoming a major factor of environmental pollution. Further, the persistence of some of these compounds in plant-derived foods (Lyndon and Darlington, 1998; Jame et al., 1999) may have a detrimental impact on both animal and human health. Nevertheless, since most of the research has focused on the effectiveness of herbicides on weeds and/or the secondary effects on crop yield, the toxicity of such substances on non-target species and the subsequent plant responses, particularly those at the molecular level, are topics that are poorly investigated. Flumioxazin (fmx) is among the newly synthesized molecules recently authorized in vineyards to control a broad spectrum of adventitious grasses and weeds (Tomlin, 2000). This soil-applied herbicide is known to be a peroxidizing agent, through the inhibition of protoporphyrinogen IX oxidase (protox IX), a key enzyme involved in chlorophyll biosynthesis (Lee and Duke, 1994). In the present study, two-dimensional (2-D) gel electrophoresis coupled to mass spectrometry (MS) analysis has been used to address the in vivo proteomic changes in non-target grapevine (Vitis vinifera L.) plantlets exposed to sublethal doses of fmx. These findings provide a framework for future investigations in order to highlight the biological impact of a herbicide widely used in viticulture, as well as to study the role of the stress-induced proteins in the fmx response pathways.
Materials and methods
Plant material and chemicals
Six-week-old grapevine plantlets were used for all the experiments. Microcuttings of Vitis vinifera L. cv. Chardonnay (clone 7535) were grown in 2.5 cm diameter glass tubes containing approximately 15 ml of MM culture medium (Martin et al., 1987) under controlled environmental conditions at 26 °C, 80–85% relative humidity, a photon flux density of 75 μmol m−1 s−1, and a 16/8 h photoperiod.
All the chemicals used had purity greater than 99%. Carrier ampholytes for IEF were purchased from Amersham-Pharmacia Biotech (Uppsala, Sweden), whereas 2-D standards were obtained from Bio-Rad (Hercules, CA, USA). All other chemicals were from Sigma (St Louis, MO, USA).
Herbicide treatments and sampling
In vitro-cultured grapevine plantlets were transferred onto fresh MM media containing either 0 (control) or 10 μM fmx. The culture medium was supplemented with PLEDGE (Cyanamid Agro, France), a water-soluble powder consisting of 50% (w/w) fmx, in order to obtain the desired concentration. At that concentration of herbicide, the grapevine plantlets were highly stressed, but they were also able to survive and continue to grow normally after an adaptation period of about 3 weeks. Control and fmx-treated plantlets (five of each) were sampled after 1, 2, 4, 6, and 21 d after transfer to the new culture medium. Sampling was carried out at approximately the same time of day in order to avoid circadian metabolic fluctuations. Leaves from the second position to the top, shoots (including petioles) and roots (previously rinsed with distilled water) were dissected and processed as described below.
Sample preparation and protein determination
Leaves, shoots and roots samples were homogenized to a very fine powder in a liquid nitrogen-precooled mortar by using a pestle. For each, approximately 0.1 g of the resulting homogenate was put into a 1.5 ml tube with 1 ml of a lysis buffer consisting of 7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 3% (w/v) SDS, 60 mM DTT, 0.5% (v/v) Pharmalyte ampholytes (pH 3–10), and 0.01% (w/v) bromophenol blue, and then suspended by vigorous shaking. In order to remove excess phenols, 5% (w/v) polyvinylpolypyrrolidone (PVPP) was added to the mixture at this step. Proteins were precipitated in 10 vols of 20% (w/v) TCA and 0.2% (w/v) DTT in chilled acetone at −20 °C for 1 h. Precipitates were resuspended in 0.5 ml of a solubilization solution (a modified lysis buffer, lacking SDS), centrifuged at 40 000 g for 60 min at 4 °C to remove all insoluble particulates, and stored at −80 °C until use. Total protein content was estimated for each sample according to the method described by Bradford (1976), using BSA as the standard and the DC reagent (Bio-Rad).
Protein expression profiling
For analytical separations, samples containing approximately 75 μg of total protein were diluted to a final volume of 350 μl in solubilization buffer, and subsequently applied by in-gel rehydration at 30 V for 10 h on to dried polyacrylamide gels (Immobiline DryStrip [pH 3–10] NL, Amersham-Pharmacia Biotech). IEF was conducted at 20 °C in an IPGPhor apparatus (Amersham-Pharmacia Biotech) as follows: 300 V and 1000 V for 1 h each followed by a linear increase from 1000 V to 8000 V, and finally 8000 V to give a total of 70 kVh. After reduction and alkylation steps (Görg et al., 1988), focused gels were placed on top of vertical slabs of acrylamide (12% T, 2.6% C). The stacking gel was replaced by a layer of 1% (w/v) agarose, 0.15 M Bis-Tris/0.1 M HCl and 0.2% (w/v) SDS. Electrophoretic migration along the second dimension was performed using the Laemmli buffer system (Laemmli, 1970) at 10 °C in a Protean II xi Cell (Bio-Rad) at 20 mA per gel for 1 h, followed by 40 mA per gel for 3 h. Reproducibility of 2-DE protein profiles was confirmed by first carrying out two independent time-course experiments (i.e. control versus herbicide exposure), and second by running each protein sample in duplicate. A total of 120 gels (3 organs×2 treatments×5 time points×2 independent experiments×2 replicas) were run and analysed.
After completion of SDS–PAGE, the gels were fixed and silver stained according to the protocol described by Rabilloud et al. (1992). Digitized images at 84.7 μm resolution were obtained using the GS-710 scanner and Quantity One software (Bio-Rad). Computerized 2-D gel analysis, including protein detection and quantification, were performed using the Melanie II software (Bio-Rad). The apparent molecular mass was calibrated using internal markers (2-D SDS–PAGE Standards, Bio-Rad) after co-electrophoresis. Protein features were modelled as Gaussians and their relative optical densities (OD), i.e. the feature OD divided by the total OD over the whole image, were computed. For each protein, OD values obtained during each experiment were normalized to the highest OD recorded, so final OD values extended from 0 (absence of protein) to 1 (highest amount of protein). Relative quantitative analyses were only performed on selected well-resolved and high quality gel spots. To simplify the calculations, the best quality gel (replica) of each experiment was chosen, and the mean value (n=2) was computed for each data point (organ×treatment×time point). The standard deviation of the analysis ranged from 0 to 33.94%. Finally, the induction/repression index for each spot was calculated as the ratio of protein expression in herbicide-treated plantlets versus control plantlets.
Preparative 2-D experiments were conducted as above with some modifications: (i) approximately 750 μg of total protein per sample was loaded onto isoelectrofocusing strips; (ii) for the first dimension, gels were run at 8000 V for a total of 140 kVh; (iii) the resulting 2-D gels were stained with Coomassie Brilliant Blue G-250 according to Anderson et al. (1991).
Mass spectrometry analysis and data interpretation
Only proteins with an induction/repression index higher than 1.8 or lower than 0.5 were sequenced by mass spectrometry. In situ digestion of protein spots was performed using the MassPREP Station (Micromass, Manchester, UK). Selected gel plugs were excised from preparative gels and washed three times in a mixture containing 25 mM NH4HCO3:ACN (1:1, v/v). The cysteine residues were reduced by 50 μl of 10 mM DTT at 57 °C and alkylated by 50 μl of 55 mM iodoacetamide at room temperature. After gel dehydration with ACN, proteins were digested overnight at room temperature in 15 μl of a solution containing 12.5 ng μl−1 of a modified porcine trypsin (Promega, Madison, WI, USA) prepared in 25 mM NH4HCO3. Finally, a double extraction was performed, first with 60% (v/v) ACN in 5% (v/v) formic acid, and subsequently with 100% (v/v) ACN. Nano-LC-MS/MS analysis of the resulting tryptic peptides was performed using a CapLC capillary LC system (Micromass) coupled to a hybrid quadrupole orthogonal acceleration time-of-flight tandem mass spectrometer (Q-TOF II, Micromass). Chromatographic separations were conducted on a Pepmap™ C18, 75 μm i.d.×15 cm length, reverse-phase (RP) capillary column (LC Packings, Sunnyvale, CA, USA) with a flow rate of 200 nl min−1, accomplished by a pre-column split. An external calibration was performed using a 2 pmol μl−1 GFP ([Glu1]-Fibrinopeptide B) solution. Mass data acquisition was piloted by MassLynx 4 software (Micromass) using automatic switching between MS and MS/MS modes.
Classical protein database searches were performed on a local Mascot™ (Matrix Science, London, UK) server. To be accepted for the identification, an error of less than 100 p.p.m. on the parent ion mass was tolerated and the sequences of the peptides were manually checked. One missed cleavage per peptide was allowed and some modifications were taken into account: carbamidomethylation for Cys, and oxidation for Met. In addition, the searches were performed without constraining protein Mr and pI, and without any taxonomic specifications. These searches did not always lead to a positive identification since the V. vinifera genome has not yet been sequenced. In such cases, the use of a de novo sequencing approach was necessary for a successful identification. For this purpose, the interpretation of the MS/MS spectra was performed with the PepSeq tool from the MassLynx 4 (Micromass) software, as well as the PEAKS Studio software (Bioinformatics Solutions, Waterloo, Canada). The resulting peptide sequences were submitted to the BLAST program provided at the EMBL site (http://dove.embl-heidelberg.de/Blast2/msblast.html) in order to identify them by homology with proteins present in the databases.
Results
Proteome changes in grapevine tissues after fmx exposure
High-resolution 2-D maps displaying up to 2000 spots were visualized from root samples. Figures 1A and B present the 2-D protein patterns obtained from control and fmx-treated root samples after 24 h of treatment, respectively. Eleven proteins showed an altered expression pattern following fmx treatments (Table 1). The number of up-regulated spots largely exceeded that of down-regulated ones. In this way, the herbicide induced the appearance of seven new proteins, whereas two spots disappeared. Concomitantly, two other prominent protein spots exhibited a significant decrease in their intensity levels after the addition of fmx. Figure 2 shows induction or repression kinetics of some of these proteins. Indeed, most of selected proteins exhibited significant symptoms of change in their expression patterns as soon as 24 h after the treatments were initiated (Table 1). Variations of protein expression still remained significant after 3 weeks of treatment except for GRP10 and GRP11, which were detectable during the first week only (Table 1).
Differentially expressed proteins in grapevine tissues following fmx exposure, revealed by expression profiling after IEF/SDS–PAGE and silver staining. Separated proteins from non-treated roots at 24 h (A), shoots at 48 h (C), and leaves at 21 d (E) are compared with protein profiles resulting from stressed roots at 24 h (B), shoots at 48 h (D), and leaves at 21 d (F) after the addition of 10 μM flumioxazin to the culture medium. Location of newly synthesized (circles) and lost (squares) proteins in the 2-D gel, as well as those spots whose quantity significantly increased (upward pointed triangle) or decreased (downward pointed triangle), is indicated. Marked proteins are named in accordance with Table 1 and were identified by nano-LC-MS/MS. The numbers at the top of the gel A denote the pH gradient in the first dimension, while the molecular masses of the 2-D standards are displayed on the left. Abbreviations: GRP, Grapevine Root Protein; GSP, Grapevine Shoot Protein; GLP, Grapevine Leaf Protein.
Expression changes of some grapevine root proteins after exposure to the herbicide flumioxazin. (A, C, E) Non-treated control plantlets. (B, D, F) Grapevine plantlets grown in the presence of 10 μM fmx. Control and fmx-treated roots were sampled after 1, 2, 4, 6, and 21 d of exposure to the herbicide. See Fig. 1 for the identification of the corresponding gel regions. Abbreviation: GRP, Grapevine Root Protein.
Time-course expression analysis of fmx-responsive proteins in grapevine tissues
Spota . | 1 d . | . | . | 2 d . | . | . | 4 d . | . | . | 6 d . | . | . | 21 d . | . | . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | Cb . | fmxc . | Changed . | C . | fmx . | Change . | C . | fmx . | Change . | C . | fmx . | Change . | C . | fmx . | Change . | ||||||||||
| GRP1 | 0.00 | 0.76 | + | 0.00 | 1.00 | + | 0.00 | 0.78 | + | 0.00 | 0.35 | + | 0.00 | 0.18 | + | ||||||||||
| GRP2 | 0.81 | 0.85 | 1.05 | 0.60 | 0.00 | − | 1.00 | 0.00 | − | 0.70 | 0.00 | − | 0.35 | 0.00 | − | ||||||||||
| GRP3 | 0.00 | 0.41 | + | 0.00 | 1.00 | + | 0.00 | 0.67 | + | 0.00 | 0.26 | + | 0.00 | 0.17 | + | ||||||||||
| GRP4 | 1.00 | 0.62 | 0.62 | 0.75 | 0.00 | − | 0.71 | 0.00 | − | 0.57 | 0.00 | − | 0.22 | 0.00 | − | ||||||||||
| GRP5 | 0.84 | 0.70 | 0.83 | 0.84 | 0.41 | 0.49 | 0.89 | 0.29 | 0.33 | 1.00 | 0.00 | − | 0.76 | 0.00 | − | ||||||||||
| GRP6 | 0.00 | 0.47 | + | 0.00 | 1.00 | + | 0.00 | 0.60 | + | 0.00 | 0.13 | + | 0.00 | 0.06 | + | ||||||||||
| GRP7 | 0.00 | 0.62 | + | 0.00 | 1.00 | + | 0.00 | 0.64 | + | 0.00 | 0.21 | + | 0.00 | 0.13 | + | ||||||||||
| GRP8 | 1.00 | 0.44 | 0.44 | 0.62 | 0.30 | 0.48 | 0.45 | 0.25 | 0.47 | 0.46 | 0.21 | 0.46 | 0.58 | 0.22 | 0.38 | ||||||||||
| GRP9 | 0.00 | 0.43 | + | 0.00 | 1.00 | + | 0.00 | 0.70 | + | 0.00 | 0.09 | + | 0.00 | 0.09 | + | ||||||||||
| GRP10 | 0.00 | 0.57 | + | 0.00 | 1.00 | + | 0.00 | 0.64 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GRP11 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.67 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GSP1 | 0.00 | 0.00 | = | 0.00 | 0.49 | + | 0.00 | 0.88 | + | 0.00 | 0.78 | + | 0.00 | 1.00 | + | ||||||||||
| GSP2 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.55 | + | 0.00 | 1.00 | + | ||||||||||
| GSP3 | 0.00 | 0.00 | = | 0.00 | 0.65 | + | 0.00 | 0.31 | + | 0.00 | 1.00 | + | 0.00 | 0.34 | + | ||||||||||
| GSP4 | 0.09 | 0.08 | 0.89 | 0.24 | 1.00 | 4.17 | 0.24 | 0.33 | 1.38 | 0.28 | 0.59 | 2.11 | 0.27 | 0.38 | 1.41 | ||||||||||
| GSP5 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.05 | 0.47 | 9.40 | 0.09 | 0.48 | 5.33 | 0.14 | 0.42 | 3.00 | ||||||||||
| GSP6 | 0.00 | 0.00 | = | 0.00 | 0.44 | + | 0.00 | 0.50 | + | 0.00 | 0.39 | + | 0.22 | 1.00 | 4.55 | ||||||||||
| GSP7 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.43 | + | 0.00 | 0.36 | + | 0.00 | 1.00 | + | ||||||||||
| GSP8 | 0.00 | 0.00 | = | 0.00 | 0.34 | + | 0.00 | 0.88 | + | 0.00 | 1.00 | + | 0.00 | 0.52 | + | ||||||||||
| GSP9 | 0.00 | 0.00 | = | 0.00 | 0.77 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GSP10 | 0.00 | 0.00 | = | 0.00 | 0.21 | + | 0.00 | 0.88 | + | 0.00 | 1.00 | + | 0.00 | 0.74 | + | ||||||||||
| GSP11 | 0.00 | 0.00 | = | 0.00 | 0.24 | + | 0.00 | 0.60 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | ||||||||||
| GSP12 | 0.03 | 0.03 | 1.00 | 0.22 | 0.43 | 1.95 | 0.02 | 0.95 | 47.50 | 0.08 | 1.00 | 12.50 | 0.19 | 0.32 | 1.68 | ||||||||||
| GSP13 | 0.00 | 0.00 | = | 0.00 | 0.40 | + | 0.00 | 0.76 | + | 0.00 | 1.00 | + | 0.00 | 0.96 | + | ||||||||||
| GLP1 | 0.00 | 0.48 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GLP2 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.62 | + | ||||||||||
| GLP3 | 0.00 | 1.00 | + | 0.00 | 0.34 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GLP4 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.51 | + | 0.00 | 0.16 | + | 0.00 | 1.00 | + | ||||||||||
| GLP5 | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.44 | + | 0.00 | 0.38 | + | 0.00 | 0.41 | + | ||||||||||
| GLP6 | 0.32 | 0.60 | 1.89 | 0.10 | 0.57 | 5.70 | 0.09 | 0.37 | 4.11 | 0.36 | 1.00 | 2.78 | 0.61 | 0.66 | 1.08 | ||||||||||
| GLP7 | 0.52 | 0.24 | 0.46 | 0.64 | 0.46 | 0.72 | 0.46 | 1.00 | 2.17 | 0.24 | 0.49 | 2.04 | 0.36 | 0.47 | 1.31 | ||||||||||
| GLP8 | 0.43 | 0.36 | 0.84 | 0.31 | 0.54 | 1.74 | 0.28 | 0.67 | 2.39 | 0.35 | 1.00 | 2.86 | 0.67 | 0.85 | 1.27 | ||||||||||
| GLP9 | 0.38 | 0.27 | 0.71 | 0.26 | 0.36 | 1.38 | 0.33 | 0.58 | 1.76 | 0.30 | 1.00 | 3.33 | 0.64 | 0.84 | 1.31 | ||||||||||
Spota . | 1 d . | . | . | 2 d . | . | . | 4 d . | . | . | 6 d . | . | . | 21 d . | . | . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | Cb . | fmxc . | Changed . | C . | fmx . | Change . | C . | fmx . | Change . | C . | fmx . | Change . | C . | fmx . | Change . | ||||||||||
| GRP1 | 0.00 | 0.76 | + | 0.00 | 1.00 | + | 0.00 | 0.78 | + | 0.00 | 0.35 | + | 0.00 | 0.18 | + | ||||||||||
| GRP2 | 0.81 | 0.85 | 1.05 | 0.60 | 0.00 | − | 1.00 | 0.00 | − | 0.70 | 0.00 | − | 0.35 | 0.00 | − | ||||||||||
| GRP3 | 0.00 | 0.41 | + | 0.00 | 1.00 | + | 0.00 | 0.67 | + | 0.00 | 0.26 | + | 0.00 | 0.17 | + | ||||||||||
| GRP4 | 1.00 | 0.62 | 0.62 | 0.75 | 0.00 | − | 0.71 | 0.00 | − | 0.57 | 0.00 | − | 0.22 | 0.00 | − | ||||||||||
| GRP5 | 0.84 | 0.70 | 0.83 | 0.84 | 0.41 | 0.49 | 0.89 | 0.29 | 0.33 | 1.00 | 0.00 | − | 0.76 | 0.00 | − | ||||||||||
| GRP6 | 0.00 | 0.47 | + | 0.00 | 1.00 | + | 0.00 | 0.60 | + | 0.00 | 0.13 | + | 0.00 | 0.06 | + | ||||||||||
| GRP7 | 0.00 | 0.62 | + | 0.00 | 1.00 | + | 0.00 | 0.64 | + | 0.00 | 0.21 | + | 0.00 | 0.13 | + | ||||||||||
| GRP8 | 1.00 | 0.44 | 0.44 | 0.62 | 0.30 | 0.48 | 0.45 | 0.25 | 0.47 | 0.46 | 0.21 | 0.46 | 0.58 | 0.22 | 0.38 | ||||||||||
| GRP9 | 0.00 | 0.43 | + | 0.00 | 1.00 | + | 0.00 | 0.70 | + | 0.00 | 0.09 | + | 0.00 | 0.09 | + | ||||||||||
| GRP10 | 0.00 | 0.57 | + | 0.00 | 1.00 | + | 0.00 | 0.64 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GRP11 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.67 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GSP1 | 0.00 | 0.00 | = | 0.00 | 0.49 | + | 0.00 | 0.88 | + | 0.00 | 0.78 | + | 0.00 | 1.00 | + | ||||||||||
| GSP2 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.55 | + | 0.00 | 1.00 | + | ||||||||||
| GSP3 | 0.00 | 0.00 | = | 0.00 | 0.65 | + | 0.00 | 0.31 | + | 0.00 | 1.00 | + | 0.00 | 0.34 | + | ||||||||||
| GSP4 | 0.09 | 0.08 | 0.89 | 0.24 | 1.00 | 4.17 | 0.24 | 0.33 | 1.38 | 0.28 | 0.59 | 2.11 | 0.27 | 0.38 | 1.41 | ||||||||||
| GSP5 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.05 | 0.47 | 9.40 | 0.09 | 0.48 | 5.33 | 0.14 | 0.42 | 3.00 | ||||||||||
| GSP6 | 0.00 | 0.00 | = | 0.00 | 0.44 | + | 0.00 | 0.50 | + | 0.00 | 0.39 | + | 0.22 | 1.00 | 4.55 | ||||||||||
| GSP7 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.43 | + | 0.00 | 0.36 | + | 0.00 | 1.00 | + | ||||||||||
| GSP8 | 0.00 | 0.00 | = | 0.00 | 0.34 | + | 0.00 | 0.88 | + | 0.00 | 1.00 | + | 0.00 | 0.52 | + | ||||||||||
| GSP9 | 0.00 | 0.00 | = | 0.00 | 0.77 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GSP10 | 0.00 | 0.00 | = | 0.00 | 0.21 | + | 0.00 | 0.88 | + | 0.00 | 1.00 | + | 0.00 | 0.74 | + | ||||||||||
| GSP11 | 0.00 | 0.00 | = | 0.00 | 0.24 | + | 0.00 | 0.60 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | ||||||||||
| GSP12 | 0.03 | 0.03 | 1.00 | 0.22 | 0.43 | 1.95 | 0.02 | 0.95 | 47.50 | 0.08 | 1.00 | 12.50 | 0.19 | 0.32 | 1.68 | ||||||||||
| GSP13 | 0.00 | 0.00 | = | 0.00 | 0.40 | + | 0.00 | 0.76 | + | 0.00 | 1.00 | + | 0.00 | 0.96 | + | ||||||||||
| GLP1 | 0.00 | 0.48 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GLP2 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.62 | + | ||||||||||
| GLP3 | 0.00 | 1.00 | + | 0.00 | 0.34 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GLP4 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.51 | + | 0.00 | 0.16 | + | 0.00 | 1.00 | + | ||||||||||
| GLP5 | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.44 | + | 0.00 | 0.38 | + | 0.00 | 0.41 | + | ||||||||||
| GLP6 | 0.32 | 0.60 | 1.89 | 0.10 | 0.57 | 5.70 | 0.09 | 0.37 | 4.11 | 0.36 | 1.00 | 2.78 | 0.61 | 0.66 | 1.08 | ||||||||||
| GLP7 | 0.52 | 0.24 | 0.46 | 0.64 | 0.46 | 0.72 | 0.46 | 1.00 | 2.17 | 0.24 | 0.49 | 2.04 | 0.36 | 0.47 | 1.31 | ||||||||||
| GLP8 | 0.43 | 0.36 | 0.84 | 0.31 | 0.54 | 1.74 | 0.28 | 0.67 | 2.39 | 0.35 | 1.00 | 2.86 | 0.67 | 0.85 | 1.27 | ||||||||||
| GLP9 | 0.38 | 0.27 | 0.71 | 0.26 | 0.36 | 1.38 | 0.33 | 0.58 | 1.76 | 0.30 | 1.00 | 3.33 | 0.64 | 0.84 | 1.31 | ||||||||||
Spots are named according to Fig. 1.
Numeric values denote the relative optical density (OD) for each protein, i.e. the feature OD divided by the total OD over the whole image, normalized to the highest value of the row.
Ratio of expression level in fmx-treated plantlets over controls. Values higher than 1.8 or lower than 0.5 were considered significant (in bold). In those cases where the ratio fmx/C could not be calculated, a new synthesis (+) or suppression (−) effect is indicated. No significant changes in protein levels are shown by the symbol =. Abbreviations: C, control plantlets; fmx, flumioxazin-treated plantlets; GRP, Grapevine Root Protein; GSP, Grapevine Shoot Protein; GLP, Grapevine Leaf Protein.
Time-course expression analysis of fmx-responsive proteins in grapevine tissues
Spota . | 1 d . | . | . | 2 d . | . | . | 4 d . | . | . | 6 d . | . | . | 21 d . | . | . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | Cb . | fmxc . | Changed . | C . | fmx . | Change . | C . | fmx . | Change . | C . | fmx . | Change . | C . | fmx . | Change . | ||||||||||
| GRP1 | 0.00 | 0.76 | + | 0.00 | 1.00 | + | 0.00 | 0.78 | + | 0.00 | 0.35 | + | 0.00 | 0.18 | + | ||||||||||
| GRP2 | 0.81 | 0.85 | 1.05 | 0.60 | 0.00 | − | 1.00 | 0.00 | − | 0.70 | 0.00 | − | 0.35 | 0.00 | − | ||||||||||
| GRP3 | 0.00 | 0.41 | + | 0.00 | 1.00 | + | 0.00 | 0.67 | + | 0.00 | 0.26 | + | 0.00 | 0.17 | + | ||||||||||
| GRP4 | 1.00 | 0.62 | 0.62 | 0.75 | 0.00 | − | 0.71 | 0.00 | − | 0.57 | 0.00 | − | 0.22 | 0.00 | − | ||||||||||
| GRP5 | 0.84 | 0.70 | 0.83 | 0.84 | 0.41 | 0.49 | 0.89 | 0.29 | 0.33 | 1.00 | 0.00 | − | 0.76 | 0.00 | − | ||||||||||
| GRP6 | 0.00 | 0.47 | + | 0.00 | 1.00 | + | 0.00 | 0.60 | + | 0.00 | 0.13 | + | 0.00 | 0.06 | + | ||||||||||
| GRP7 | 0.00 | 0.62 | + | 0.00 | 1.00 | + | 0.00 | 0.64 | + | 0.00 | 0.21 | + | 0.00 | 0.13 | + | ||||||||||
| GRP8 | 1.00 | 0.44 | 0.44 | 0.62 | 0.30 | 0.48 | 0.45 | 0.25 | 0.47 | 0.46 | 0.21 | 0.46 | 0.58 | 0.22 | 0.38 | ||||||||||
| GRP9 | 0.00 | 0.43 | + | 0.00 | 1.00 | + | 0.00 | 0.70 | + | 0.00 | 0.09 | + | 0.00 | 0.09 | + | ||||||||||
| GRP10 | 0.00 | 0.57 | + | 0.00 | 1.00 | + | 0.00 | 0.64 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GRP11 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.67 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GSP1 | 0.00 | 0.00 | = | 0.00 | 0.49 | + | 0.00 | 0.88 | + | 0.00 | 0.78 | + | 0.00 | 1.00 | + | ||||||||||
| GSP2 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.55 | + | 0.00 | 1.00 | + | ||||||||||
| GSP3 | 0.00 | 0.00 | = | 0.00 | 0.65 | + | 0.00 | 0.31 | + | 0.00 | 1.00 | + | 0.00 | 0.34 | + | ||||||||||
| GSP4 | 0.09 | 0.08 | 0.89 | 0.24 | 1.00 | 4.17 | 0.24 | 0.33 | 1.38 | 0.28 | 0.59 | 2.11 | 0.27 | 0.38 | 1.41 | ||||||||||
| GSP5 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.05 | 0.47 | 9.40 | 0.09 | 0.48 | 5.33 | 0.14 | 0.42 | 3.00 | ||||||||||
| GSP6 | 0.00 | 0.00 | = | 0.00 | 0.44 | + | 0.00 | 0.50 | + | 0.00 | 0.39 | + | 0.22 | 1.00 | 4.55 | ||||||||||
| GSP7 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.43 | + | 0.00 | 0.36 | + | 0.00 | 1.00 | + | ||||||||||
| GSP8 | 0.00 | 0.00 | = | 0.00 | 0.34 | + | 0.00 | 0.88 | + | 0.00 | 1.00 | + | 0.00 | 0.52 | + | ||||||||||
| GSP9 | 0.00 | 0.00 | = | 0.00 | 0.77 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GSP10 | 0.00 | 0.00 | = | 0.00 | 0.21 | + | 0.00 | 0.88 | + | 0.00 | 1.00 | + | 0.00 | 0.74 | + | ||||||||||
| GSP11 | 0.00 | 0.00 | = | 0.00 | 0.24 | + | 0.00 | 0.60 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | ||||||||||
| GSP12 | 0.03 | 0.03 | 1.00 | 0.22 | 0.43 | 1.95 | 0.02 | 0.95 | 47.50 | 0.08 | 1.00 | 12.50 | 0.19 | 0.32 | 1.68 | ||||||||||
| GSP13 | 0.00 | 0.00 | = | 0.00 | 0.40 | + | 0.00 | 0.76 | + | 0.00 | 1.00 | + | 0.00 | 0.96 | + | ||||||||||
| GLP1 | 0.00 | 0.48 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GLP2 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.62 | + | ||||||||||
| GLP3 | 0.00 | 1.00 | + | 0.00 | 0.34 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GLP4 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.51 | + | 0.00 | 0.16 | + | 0.00 | 1.00 | + | ||||||||||
| GLP5 | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.44 | + | 0.00 | 0.38 | + | 0.00 | 0.41 | + | ||||||||||
| GLP6 | 0.32 | 0.60 | 1.89 | 0.10 | 0.57 | 5.70 | 0.09 | 0.37 | 4.11 | 0.36 | 1.00 | 2.78 | 0.61 | 0.66 | 1.08 | ||||||||||
| GLP7 | 0.52 | 0.24 | 0.46 | 0.64 | 0.46 | 0.72 | 0.46 | 1.00 | 2.17 | 0.24 | 0.49 | 2.04 | 0.36 | 0.47 | 1.31 | ||||||||||
| GLP8 | 0.43 | 0.36 | 0.84 | 0.31 | 0.54 | 1.74 | 0.28 | 0.67 | 2.39 | 0.35 | 1.00 | 2.86 | 0.67 | 0.85 | 1.27 | ||||||||||
| GLP9 | 0.38 | 0.27 | 0.71 | 0.26 | 0.36 | 1.38 | 0.33 | 0.58 | 1.76 | 0.30 | 1.00 | 3.33 | 0.64 | 0.84 | 1.31 | ||||||||||
Spota . | 1 d . | . | . | 2 d . | . | . | 4 d . | . | . | 6 d . | . | . | 21 d . | . | . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | Cb . | fmxc . | Changed . | C . | fmx . | Change . | C . | fmx . | Change . | C . | fmx . | Change . | C . | fmx . | Change . | ||||||||||
| GRP1 | 0.00 | 0.76 | + | 0.00 | 1.00 | + | 0.00 | 0.78 | + | 0.00 | 0.35 | + | 0.00 | 0.18 | + | ||||||||||
| GRP2 | 0.81 | 0.85 | 1.05 | 0.60 | 0.00 | − | 1.00 | 0.00 | − | 0.70 | 0.00 | − | 0.35 | 0.00 | − | ||||||||||
| GRP3 | 0.00 | 0.41 | + | 0.00 | 1.00 | + | 0.00 | 0.67 | + | 0.00 | 0.26 | + | 0.00 | 0.17 | + | ||||||||||
| GRP4 | 1.00 | 0.62 | 0.62 | 0.75 | 0.00 | − | 0.71 | 0.00 | − | 0.57 | 0.00 | − | 0.22 | 0.00 | − | ||||||||||
| GRP5 | 0.84 | 0.70 | 0.83 | 0.84 | 0.41 | 0.49 | 0.89 | 0.29 | 0.33 | 1.00 | 0.00 | − | 0.76 | 0.00 | − | ||||||||||
| GRP6 | 0.00 | 0.47 | + | 0.00 | 1.00 | + | 0.00 | 0.60 | + | 0.00 | 0.13 | + | 0.00 | 0.06 | + | ||||||||||
| GRP7 | 0.00 | 0.62 | + | 0.00 | 1.00 | + | 0.00 | 0.64 | + | 0.00 | 0.21 | + | 0.00 | 0.13 | + | ||||||||||
| GRP8 | 1.00 | 0.44 | 0.44 | 0.62 | 0.30 | 0.48 | 0.45 | 0.25 | 0.47 | 0.46 | 0.21 | 0.46 | 0.58 | 0.22 | 0.38 | ||||||||||
| GRP9 | 0.00 | 0.43 | + | 0.00 | 1.00 | + | 0.00 | 0.70 | + | 0.00 | 0.09 | + | 0.00 | 0.09 | + | ||||||||||
| GRP10 | 0.00 | 0.57 | + | 0.00 | 1.00 | + | 0.00 | 0.64 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GRP11 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.67 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GSP1 | 0.00 | 0.00 | = | 0.00 | 0.49 | + | 0.00 | 0.88 | + | 0.00 | 0.78 | + | 0.00 | 1.00 | + | ||||||||||
| GSP2 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.55 | + | 0.00 | 1.00 | + | ||||||||||
| GSP3 | 0.00 | 0.00 | = | 0.00 | 0.65 | + | 0.00 | 0.31 | + | 0.00 | 1.00 | + | 0.00 | 0.34 | + | ||||||||||
| GSP4 | 0.09 | 0.08 | 0.89 | 0.24 | 1.00 | 4.17 | 0.24 | 0.33 | 1.38 | 0.28 | 0.59 | 2.11 | 0.27 | 0.38 | 1.41 | ||||||||||
| GSP5 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.05 | 0.47 | 9.40 | 0.09 | 0.48 | 5.33 | 0.14 | 0.42 | 3.00 | ||||||||||
| GSP6 | 0.00 | 0.00 | = | 0.00 | 0.44 | + | 0.00 | 0.50 | + | 0.00 | 0.39 | + | 0.22 | 1.00 | 4.55 | ||||||||||
| GSP7 | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.43 | + | 0.00 | 0.36 | + | 0.00 | 1.00 | + | ||||||||||
| GSP8 | 0.00 | 0.00 | = | 0.00 | 0.34 | + | 0.00 | 0.88 | + | 0.00 | 1.00 | + | 0.00 | 0.52 | + | ||||||||||
| GSP9 | 0.00 | 0.00 | = | 0.00 | 0.77 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GSP10 | 0.00 | 0.00 | = | 0.00 | 0.21 | + | 0.00 | 0.88 | + | 0.00 | 1.00 | + | 0.00 | 0.74 | + | ||||||||||
| GSP11 | 0.00 | 0.00 | = | 0.00 | 0.24 | + | 0.00 | 0.60 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | ||||||||||
| GSP12 | 0.03 | 0.03 | 1.00 | 0.22 | 0.43 | 1.95 | 0.02 | 0.95 | 47.50 | 0.08 | 1.00 | 12.50 | 0.19 | 0.32 | 1.68 | ||||||||||
| GSP13 | 0.00 | 0.00 | = | 0.00 | 0.40 | + | 0.00 | 0.76 | + | 0.00 | 1.00 | + | 0.00 | 0.96 | + | ||||||||||
| GLP1 | 0.00 | 0.48 | + | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GLP2 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 1.00 | + | 0.00 | 0.62 | + | ||||||||||
| GLP3 | 0.00 | 1.00 | + | 0.00 | 0.34 | + | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.00 | = | ||||||||||
| GLP4 | 0.00 | 0.00 | = | 0.00 | 0.00 | = | 0.00 | 0.51 | + | 0.00 | 0.16 | + | 0.00 | 1.00 | + | ||||||||||
| GLP5 | 0.00 | 1.00 | + | 0.00 | 0.00 | = | 0.00 | 0.44 | + | 0.00 | 0.38 | + | 0.00 | 0.41 | + | ||||||||||
| GLP6 | 0.32 | 0.60 | 1.89 | 0.10 | 0.57 | 5.70 | 0.09 | 0.37 | 4.11 | 0.36 | 1.00 | 2.78 | 0.61 | 0.66 | 1.08 | ||||||||||
| GLP7 | 0.52 | 0.24 | 0.46 | 0.64 | 0.46 | 0.72 | 0.46 | 1.00 | 2.17 | 0.24 | 0.49 | 2.04 | 0.36 | 0.47 | 1.31 | ||||||||||
| GLP8 | 0.43 | 0.36 | 0.84 | 0.31 | 0.54 | 1.74 | 0.28 | 0.67 | 2.39 | 0.35 | 1.00 | 2.86 | 0.67 | 0.85 | 1.27 | ||||||||||
| GLP9 | 0.38 | 0.27 | 0.71 | 0.26 | 0.36 | 1.38 | 0.33 | 0.58 | 1.76 | 0.30 | 1.00 | 3.33 | 0.64 | 0.84 | 1.31 | ||||||||||
Spots are named according to Fig. 1.
Numeric values denote the relative optical density (OD) for each protein, i.e. the feature OD divided by the total OD over the whole image, normalized to the highest value of the row.
Ratio of expression level in fmx-treated plantlets over controls. Values higher than 1.8 or lower than 0.5 were considered significant (in bold). In those cases where the ratio fmx/C could not be calculated, a new synthesis (+) or suppression (−) effect is indicated. No significant changes in protein levels are shown by the symbol =. Abbreviations: C, control plantlets; fmx, flumioxazin-treated plantlets; GRP, Grapevine Root Protein; GSP, Grapevine Shoot Protein; GLP, Grapevine Leaf Protein.
Shoot protein analysis also resulted in complex 2-D gels consisting of about 1500 proteins. A total of 13 protein spots displayed significant changes in their expression levels in fmx-treated plantlets compared with controls (Table 1). Figure 1C and D denote differentially expressed proteins in grapevine shoots following fmx exposure. Looking individually, nine protein spots were induced from non-detectable levels upon treatments, while four other pre-existing spots were significantly amplified in amount with regard to controls (Table 1). Conversely to roots, no shoot protein was reported to disappear as a result of the herbicide stress. Figure 3 shows the accumulation of several proteins in shoot tissues during time-course experiments. It should also be noted that a delay in the response of shoot proteins was observed compared with changes in the root proteome. Indeed, the first noticeable variations in the protein pattern of shoots started 48 h after the transfer of plantlets to the herbicide-containing medium (Table 1). In addition, GSP2 only reached detectable levels after 6 d of treatments and GSP4 showed two peaks of intensity at days 2 and 6. Similarly to roots, differences were still relevant for most of the proteins 3 weeks after the onset of experiments, indicating a long-term action of the herbicide in the shoots as well (Table 1).
Detailed portions of silver-stained 2-D gels obtained from control grapevine shoots (A, B) compared with shoots treated with 10 μM fmx (B, D), showing the transient accumulation of several proteins. Control and fmx-treated shoots were sampled after 1, 2, 4, 6, and 21 d of exposure to the herbicide. See Fig. 1 for the identification of the corresponding gel regions. Abbreviation: GSP, Grapevine Shoot Protein.
Overall, more than 2000 GLP spots were identified by digital image analysis in all the gels analysed. Although the resulting protein patterns corresponding to control and fmx-treated samples looked rather similar at first sight, nine spots varied their expression due to the action of the herbicide (Table 1). Figure 1E and F show the positions of differentially expressed proteins in leaves of plantlets harvested 3 weeks after herbicide exposure. All changes in leaves concerned either proteins induced from non-detectable levels or pre-existing proteins that increased significantly (Table 1). Figure 4 presents variations in expression levels of some proteins in fmx-stressed leaves with regard to the controls. The response timing in leaves was found to be remarkably more variable as compared with roots and shoots. Indeed, five proteins were rapidly induced proteins since the first variations in their expression levels were detected 24 h after the onset of experiments, whereas the remaining proteins presented a more or less important delay in their responses (Table 1). Strikingly, the spot identified as GLP7 was initially repressed and increased its expression at the end of the first week of treatment (Table 1). Another remarkable point was that the effects on leaves seemed to be attenuated since only three proteins were found to fluctuate during the whole experiment.
Some protein changes in fmx-stressed leaves (B, D) compared with non-treated controls (A, C), during time-course experiments. Control and fmx-treated leaves were sampled after 1, 2, 4, 6, and 21 d of exposure to the herbicide. See Fig. 1 for the identification of gel regions. Abbreviation: GLP, Grapevine Leaf Protein.
The expression changes detected for all these proteins were reproducible among replicas and between the two independent experiments carried out.
Identification of fmx-responsive proteins by mass spectrometry analysis
Table 2 lists those fmx-responsive proteins identified by nano-LC-MS/MS. As the V. vinifera genome has not yet been sequenced, a de novo sequencing strategy was necessary to lead to successful protein identifications. This strategy implies the individual treatment of the peptide fragmentation spectra to deduce a complete or partial peptide sequence tag. The peptide sequences determined by this way are then submitted to a BLAST program that will allow the identification of the protein function by homology with proteins that are present in the databases. In roots, mass spectrometry analysis led to the identification of two enzymes, 2,3-biphosphoglycerate-independent phosphoglycerate mutase (PGM-i) and UTP-glucose pyrophosphorylase (UGPase), both involved in carbon metabolism, which showed best coverage scores of 15% and 16%, respectively. Moreover, spot GRP4 was identified as an orcinol O-methyltranferase (OOMT), whereas GRP5 showed homology with a D-protein, although in both cases matched peptides covered a low percentage of the protein sequence (3% and 10%, respectively). Interestingly, spots GRP6, GRP7, GRP9, GRP10, and GRP11, the most potently induced spots, were homologous to a pathogenesis-related protein from V. vinifera that belongs to Class 10 (PRP-10). All these proteins presented high coverage scores ranging between 54% and 64%. GRP7 also covered about 6% of the amino acid sequence of an adenosine kinase 1 (ADK1), although its expected Mr was significantly higher than the Mr calculated for the protein spot (Table 2), suggesting that a breakdown product of the enzyme has also co-migrated with PRP-10. Spots called GRP1 and GRP8 could not be identified because of the low signal in the MS analysis.
List of grapevine fmx-responsive proteins identified by nano-LC-MS/MS after IEF/SDS–PAGE
Spota . | Tissue . | Massesb . | Identified protein (species) . | Accession no.c . | Matching peptidesd . | %e . |
|---|---|---|---|---|---|---|
| GRP1 | Root | 61804/- | No identification | – | – | – |
| GRP2 | Root | 61804/53394 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (Prunus dulcis) | O24246 | LDQLQLLLK-GVDAQIASGGGR-AVEIAEK-LYEGEGFK | 15 |
| GRP3 | Root | 54911/51887 | UTP-glucose-1P-uridyltransferase (Arabidopsis thaliana) | P57751 | LVEADALK-LVQLETAAGAAIR-SGFINLVSR | 16 |
| GRP4 | Root | 37635/41278 | Orcinol O-methyltransferase (Rosa hybrid) | Q8L5K8 | VIIIDMMMENQK-CTVLDLPHVRAD-FNEAMASDAR-VDVGGGTG- DAGFSGYK-EEGYVLTHAS | 3 |
| GRP5 | Root | 34801/34021 | D-protein (Hordeum vulgare) | Q8VWY8 | SSHALALVGQK-LLSSGDAGPPHR-AYVFGGELTPR | 10 |
| GRP6 | Root | 20651/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 57 |
| GRP7 | Root | 20673/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | GGKEDALATFK-EDALATFK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 58 |
| 20673/37836 | Adenosin kinase 1 (Arabidopsis thaliana)f | Q9SF85 | SLIANLSAAGGCYK-LNNAILAEDK- AGCYASHVPEQR-SFPVLLLPK | 6 | ||
| GRP8 | Root | 18460/- | No identification | – | – | – |
| GRP9 | Root | 19935/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-AAVLDADNLIPK- AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 54 |
| GRP10 | Root | 20710/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | VDAIDKEK-AAVLDADNLIPK- AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 64 |
| GRP11 | Root | 20698/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 63 |
| GSP1 | Shoot | 37590/- | No identification | – | – | – |
| GSP2 | Shoot | 32385/- | No identification | – | – | – |
| GSP3 | Shoot | 40019/39073 | Glyceraldehyde-3P-dehydrogenase (Atriplex nummularia) | P34783 | VIISAPSK-VLPALNGK-LTGMSFR- SSIFDAK-AASFNIIPSSTGAAK- LVSWYDNEWGYSSR | 25 |
| GSP4 | Shoot | 21618/17419 | Thioredoxin peroxidase 1 (Nicotiana sylvestris) | Q8S310 | HVPGFIEK-VLFGVPGAFT | 11 |
| GSP5 | Shoot | 18929/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | VVASPDGGSIYK-AAILDSDNLIPK- AIEAYVLAHPDAY | 23 |
| GSP6 | Shoot | 18605/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | GGKEDALATFK-VVASPDGGSIYK- AAILDSDNLIPK-GVFTYEQEITSSVPPAK | 32 |
| GSP7 | Shoot | 18345/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | AIEAYVLAHPDAY-AAILDSDNLIPK | 15 |
| GSP8 | Shoot | 19935/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | AIEAYVLAHPDAY-AAVLDADNLIPK- GVFTYEQEITSSVPPAK-GAEVCEEHVK | 15 |
| GSP9 | Shoot | 17211/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | AAVLDADNLIPK | 7 |
| GSP10 | Shoot | 20569/17118 | PRP-10(Vitis vinifera) | Q9FS43 | GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- GVFTYEQEITSSVPPAK | 39 |
| GSP11 | Shoot | 20626/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-AAVLDADNLIPK- AIEAYVLAHPDAY- GVFTYEQEITSSVPPAK | 26 |
| GSP12 | Shoot | 20626/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | AAILDSDNLIPK-AIEAYVLAHPDAY-GVFTYEQEITSSVPPAK | 36 |
| GSP13 | Shoot | 16883/52891 | Rubisco large subunit (fragment) (Lantana camara) | Q9THN3 | DNGLLLHIHR-FLFCAEALFK- GGLDFTK-AETGEIK | 8 |
| GLP1 | Leaf | 54767/51983 | ATP synthase beta subunit (Sparganium americanum) | Q95FJ9 | ESGVINEK-IGLFGGAGVGK-LSIFETGIK- SAPAFIQLDTK-MPNIYNALVVK- TVLIMELINNIAK- IAQIIGPVLDAVFPPGK IFNVLGEPVDNLGPVDTR | 50 |
| GLP2 | Leaf | 32590/34840 | Putative 33 kDa oxygen evolving protein of photosystem II (Oryza sativa) | Q943W1 | FCLEPTSFTVK-VPFLFTIK-GSSFLDPK- GGSTGYDNAVALPAGGR | 20 |
| 32590/34489 | Carbonic anhydrase, chloroplast precursor (Nicotiana tabacum)f | P27141 | YSRGAALEYAVLHLK-YMVFACVCSR | 8 | ||
| GLP3 | Leaf | 40890/40260 | Glycolate oxidase, perisomal (Spinacia oleracea) | Q9FS42 | AIALTVDTPR-IPVFLDGGVR | 5 |
| GLP4 | Leaf | 37334/51471 | Rubisco large chain (fragment) (Bursera inaguensis) | P28385 | IPPAYSK-TFQGPPHGIQVER-AVYECLR- GGLDFTK-FLFCAEALFK-SKAETGELK DNGLLLHLHR | 19 |
| GLP5 | Leaf | 37736/35682 | Glyceraldehyde 3-phosphate dehydrogenase (Sus scrofa) | P00355 | VGVNGMGR-LTGFSMR- QAKNLLPASTGAAK- EPSWYDNEFGYSNR | 43 |
| 37736/51471 | Rubisco large chain (fragment) (Vitis vinifera)f | Q6ZYB1 | LTYYTPEYETKPTDILAAFR-IPPAYSK- TFQGPPHGIQVER-AVYECLR- GGLDFTK-GGLDFTKDDENVNSQPFMR-FLFCAEAIFK-SQAETGEIK- GHYLNATAGTCEEMIK-DNGLLLHIHR | 19 | ||
| GLP6 | Leaf | 38759/43463 | Glycine cleavage complex T-protein (fragment) (Zea mays) | Q8W521 | LTGLGAR-TVLYDFH- LALQGPLAAPVL-DVSWHLHDER | 28 |
| GLP7 | Leaf | 27954/28858 | LHCII type III cab binding protein (Vigna radiata) | gi | 4689380 | WAMLGALGCITPEVLEK- GPLENLLDHLDNPVANNAWVYATK | 15 |
| 27954/10291 | Probable oxygen-evolving enhancer protein 2 (Vitis vinifera)f | gi | 37903240 | TADGDEGGKHQLITAAVADGK- HQLITAAVADGK-KYVESTASSFSIA | 8 | ||
| GLP8 | Leaf | 29850/27033 | Ascorbate peroxidase (Vigna unguiculata) | Q41712 | ELLSGEK-EGLLQLRSDK- GPWTSNPLLFDNSYFK-PEPPPEGR- LLAGPNK-ALLSDPAFRLPVEK | 21 |
| GLP9 | Leaf | 28646/27033 | Ascorbate peroxidase (Vigna unguiculata) | Q41712 | EDKPEPPPEGR- SGFEGPWTSNPLIFDNSYFK ELLSGEK-EGLLQLPSDK | 46 |
Spota . | Tissue . | Massesb . | Identified protein (species) . | Accession no.c . | Matching peptidesd . | %e . |
|---|---|---|---|---|---|---|
| GRP1 | Root | 61804/- | No identification | – | – | – |
| GRP2 | Root | 61804/53394 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (Prunus dulcis) | O24246 | LDQLQLLLK-GVDAQIASGGGR-AVEIAEK-LYEGEGFK | 15 |
| GRP3 | Root | 54911/51887 | UTP-glucose-1P-uridyltransferase (Arabidopsis thaliana) | P57751 | LVEADALK-LVQLETAAGAAIR-SGFINLVSR | 16 |
| GRP4 | Root | 37635/41278 | Orcinol O-methyltransferase (Rosa hybrid) | Q8L5K8 | VIIIDMMMENQK-CTVLDLPHVRAD-FNEAMASDAR-VDVGGGTG- DAGFSGYK-EEGYVLTHAS | 3 |
| GRP5 | Root | 34801/34021 | D-protein (Hordeum vulgare) | Q8VWY8 | SSHALALVGQK-LLSSGDAGPPHR-AYVFGGELTPR | 10 |
| GRP6 | Root | 20651/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 57 |
| GRP7 | Root | 20673/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | GGKEDALATFK-EDALATFK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 58 |
| 20673/37836 | Adenosin kinase 1 (Arabidopsis thaliana)f | Q9SF85 | SLIANLSAAGGCYK-LNNAILAEDK- AGCYASHVPEQR-SFPVLLLPK | 6 | ||
| GRP8 | Root | 18460/- | No identification | – | – | – |
| GRP9 | Root | 19935/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-AAVLDADNLIPK- AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 54 |
| GRP10 | Root | 20710/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | VDAIDKEK-AAVLDADNLIPK- AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 64 |
| GRP11 | Root | 20698/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 63 |
| GSP1 | Shoot | 37590/- | No identification | – | – | – |
| GSP2 | Shoot | 32385/- | No identification | – | – | – |
| GSP3 | Shoot | 40019/39073 | Glyceraldehyde-3P-dehydrogenase (Atriplex nummularia) | P34783 | VIISAPSK-VLPALNGK-LTGMSFR- SSIFDAK-AASFNIIPSSTGAAK- LVSWYDNEWGYSSR | 25 |
| GSP4 | Shoot | 21618/17419 | Thioredoxin peroxidase 1 (Nicotiana sylvestris) | Q8S310 | HVPGFIEK-VLFGVPGAFT | 11 |
| GSP5 | Shoot | 18929/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | VVASPDGGSIYK-AAILDSDNLIPK- AIEAYVLAHPDAY | 23 |
| GSP6 | Shoot | 18605/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | GGKEDALATFK-VVASPDGGSIYK- AAILDSDNLIPK-GVFTYEQEITSSVPPAK | 32 |
| GSP7 | Shoot | 18345/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | AIEAYVLAHPDAY-AAILDSDNLIPK | 15 |
| GSP8 | Shoot | 19935/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | AIEAYVLAHPDAY-AAVLDADNLIPK- GVFTYEQEITSSVPPAK-GAEVCEEHVK | 15 |
| GSP9 | Shoot | 17211/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | AAVLDADNLIPK | 7 |
| GSP10 | Shoot | 20569/17118 | PRP-10(Vitis vinifera) | Q9FS43 | GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- GVFTYEQEITSSVPPAK | 39 |
| GSP11 | Shoot | 20626/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-AAVLDADNLIPK- AIEAYVLAHPDAY- GVFTYEQEITSSVPPAK | 26 |
| GSP12 | Shoot | 20626/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | AAILDSDNLIPK-AIEAYVLAHPDAY-GVFTYEQEITSSVPPAK | 36 |
| GSP13 | Shoot | 16883/52891 | Rubisco large subunit (fragment) (Lantana camara) | Q9THN3 | DNGLLLHIHR-FLFCAEALFK- GGLDFTK-AETGEIK | 8 |
| GLP1 | Leaf | 54767/51983 | ATP synthase beta subunit (Sparganium americanum) | Q95FJ9 | ESGVINEK-IGLFGGAGVGK-LSIFETGIK- SAPAFIQLDTK-MPNIYNALVVK- TVLIMELINNIAK- IAQIIGPVLDAVFPPGK IFNVLGEPVDNLGPVDTR | 50 |
| GLP2 | Leaf | 32590/34840 | Putative 33 kDa oxygen evolving protein of photosystem II (Oryza sativa) | Q943W1 | FCLEPTSFTVK-VPFLFTIK-GSSFLDPK- GGSTGYDNAVALPAGGR | 20 |
| 32590/34489 | Carbonic anhydrase, chloroplast precursor (Nicotiana tabacum)f | P27141 | YSRGAALEYAVLHLK-YMVFACVCSR | 8 | ||
| GLP3 | Leaf | 40890/40260 | Glycolate oxidase, perisomal (Spinacia oleracea) | Q9FS42 | AIALTVDTPR-IPVFLDGGVR | 5 |
| GLP4 | Leaf | 37334/51471 | Rubisco large chain (fragment) (Bursera inaguensis) | P28385 | IPPAYSK-TFQGPPHGIQVER-AVYECLR- GGLDFTK-FLFCAEALFK-SKAETGELK DNGLLLHLHR | 19 |
| GLP5 | Leaf | 37736/35682 | Glyceraldehyde 3-phosphate dehydrogenase (Sus scrofa) | P00355 | VGVNGMGR-LTGFSMR- QAKNLLPASTGAAK- EPSWYDNEFGYSNR | 43 |
| 37736/51471 | Rubisco large chain (fragment) (Vitis vinifera)f | Q6ZYB1 | LTYYTPEYETKPTDILAAFR-IPPAYSK- TFQGPPHGIQVER-AVYECLR- GGLDFTK-GGLDFTKDDENVNSQPFMR-FLFCAEAIFK-SQAETGEIK- GHYLNATAGTCEEMIK-DNGLLLHIHR | 19 | ||
| GLP6 | Leaf | 38759/43463 | Glycine cleavage complex T-protein (fragment) (Zea mays) | Q8W521 | LTGLGAR-TVLYDFH- LALQGPLAAPVL-DVSWHLHDER | 28 |
| GLP7 | Leaf | 27954/28858 | LHCII type III cab binding protein (Vigna radiata) | gi | 4689380 | WAMLGALGCITPEVLEK- GPLENLLDHLDNPVANNAWVYATK | 15 |
| 27954/10291 | Probable oxygen-evolving enhancer protein 2 (Vitis vinifera)f | gi | 37903240 | TADGDEGGKHQLITAAVADGK- HQLITAAVADGK-KYVESTASSFSIA | 8 | ||
| GLP8 | Leaf | 29850/27033 | Ascorbate peroxidase (Vigna unguiculata) | Q41712 | ELLSGEK-EGLLQLRSDK- GPWTSNPLLFDNSYFK-PEPPPEGR- LLAGPNK-ALLSDPAFRLPVEK | 21 |
| GLP9 | Leaf | 28646/27033 | Ascorbate peroxidase (Vigna unguiculata) | Q41712 | EDKPEPPPEGR- SGFEGPWTSNPLIFDNSYFK ELLSGEK-EGLLQLPSDK | 46 |
Spots are named accordingly to Fig. 1.
Observed molecular mass determined on the gel (Daltons)/Theoretical molecular mass (Daltons).
Accession number in NCBI or SWISS-PROT databases and organism assignment after BLAST homology searches.
Peptides identified via the Mascot search engine and confirmed by de novo sequencing are indicated in regular characters, whereas peptides only identified by de novo sequencing and BLAST search are in bold and italic.
Percentage of amino acids in reference proteins covered by matching peptides from nano-LC-MS/MS analysis.
Co-migrating protein. Abbreviations: GRP, Grapevine Root Protein; GSP, Grapevine Shoot Protein; GLP, Grapevine Leaf Protein.
List of grapevine fmx-responsive proteins identified by nano-LC-MS/MS after IEF/SDS–PAGE
Spota . | Tissue . | Massesb . | Identified protein (species) . | Accession no.c . | Matching peptidesd . | %e . |
|---|---|---|---|---|---|---|
| GRP1 | Root | 61804/- | No identification | – | – | – |
| GRP2 | Root | 61804/53394 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (Prunus dulcis) | O24246 | LDQLQLLLK-GVDAQIASGGGR-AVEIAEK-LYEGEGFK | 15 |
| GRP3 | Root | 54911/51887 | UTP-glucose-1P-uridyltransferase (Arabidopsis thaliana) | P57751 | LVEADALK-LVQLETAAGAAIR-SGFINLVSR | 16 |
| GRP4 | Root | 37635/41278 | Orcinol O-methyltransferase (Rosa hybrid) | Q8L5K8 | VIIIDMMMENQK-CTVLDLPHVRAD-FNEAMASDAR-VDVGGGTG- DAGFSGYK-EEGYVLTHAS | 3 |
| GRP5 | Root | 34801/34021 | D-protein (Hordeum vulgare) | Q8VWY8 | SSHALALVGQK-LLSSGDAGPPHR-AYVFGGELTPR | 10 |
| GRP6 | Root | 20651/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 57 |
| GRP7 | Root | 20673/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | GGKEDALATFK-EDALATFK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 58 |
| 20673/37836 | Adenosin kinase 1 (Arabidopsis thaliana)f | Q9SF85 | SLIANLSAAGGCYK-LNNAILAEDK- AGCYASHVPEQR-SFPVLLLPK | 6 | ||
| GRP8 | Root | 18460/- | No identification | – | – | – |
| GRP9 | Root | 19935/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-AAVLDADNLIPK- AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 54 |
| GRP10 | Root | 20710/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | VDAIDKEK-AAVLDADNLIPK- AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 64 |
| GRP11 | Root | 20698/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 63 |
| GSP1 | Shoot | 37590/- | No identification | – | – | – |
| GSP2 | Shoot | 32385/- | No identification | – | – | – |
| GSP3 | Shoot | 40019/39073 | Glyceraldehyde-3P-dehydrogenase (Atriplex nummularia) | P34783 | VIISAPSK-VLPALNGK-LTGMSFR- SSIFDAK-AASFNIIPSSTGAAK- LVSWYDNEWGYSSR | 25 |
| GSP4 | Shoot | 21618/17419 | Thioredoxin peroxidase 1 (Nicotiana sylvestris) | Q8S310 | HVPGFIEK-VLFGVPGAFT | 11 |
| GSP5 | Shoot | 18929/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | VVASPDGGSIYK-AAILDSDNLIPK- AIEAYVLAHPDAY | 23 |
| GSP6 | Shoot | 18605/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | GGKEDALATFK-VVASPDGGSIYK- AAILDSDNLIPK-GVFTYEQEITSSVPPAK | 32 |
| GSP7 | Shoot | 18345/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | AIEAYVLAHPDAY-AAILDSDNLIPK | 15 |
| GSP8 | Shoot | 19935/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | AIEAYVLAHPDAY-AAVLDADNLIPK- GVFTYEQEITSSVPPAK-GAEVCEEHVK | 15 |
| GSP9 | Shoot | 17211/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | AAVLDADNLIPK | 7 |
| GSP10 | Shoot | 20569/17118 | PRP-10(Vitis vinifera) | Q9FS43 | GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- GVFTYEQEITSSVPPAK | 39 |
| GSP11 | Shoot | 20626/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-AAVLDADNLIPK- AIEAYVLAHPDAY- GVFTYEQEITSSVPPAK | 26 |
| GSP12 | Shoot | 20626/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | AAILDSDNLIPK-AIEAYVLAHPDAY-GVFTYEQEITSSVPPAK | 36 |
| GSP13 | Shoot | 16883/52891 | Rubisco large subunit (fragment) (Lantana camara) | Q9THN3 | DNGLLLHIHR-FLFCAEALFK- GGLDFTK-AETGEIK | 8 |
| GLP1 | Leaf | 54767/51983 | ATP synthase beta subunit (Sparganium americanum) | Q95FJ9 | ESGVINEK-IGLFGGAGVGK-LSIFETGIK- SAPAFIQLDTK-MPNIYNALVVK- TVLIMELINNIAK- IAQIIGPVLDAVFPPGK IFNVLGEPVDNLGPVDTR | 50 |
| GLP2 | Leaf | 32590/34840 | Putative 33 kDa oxygen evolving protein of photosystem II (Oryza sativa) | Q943W1 | FCLEPTSFTVK-VPFLFTIK-GSSFLDPK- GGSTGYDNAVALPAGGR | 20 |
| 32590/34489 | Carbonic anhydrase, chloroplast precursor (Nicotiana tabacum)f | P27141 | YSRGAALEYAVLHLK-YMVFACVCSR | 8 | ||
| GLP3 | Leaf | 40890/40260 | Glycolate oxidase, perisomal (Spinacia oleracea) | Q9FS42 | AIALTVDTPR-IPVFLDGGVR | 5 |
| GLP4 | Leaf | 37334/51471 | Rubisco large chain (fragment) (Bursera inaguensis) | P28385 | IPPAYSK-TFQGPPHGIQVER-AVYECLR- GGLDFTK-FLFCAEALFK-SKAETGELK DNGLLLHLHR | 19 |
| GLP5 | Leaf | 37736/35682 | Glyceraldehyde 3-phosphate dehydrogenase (Sus scrofa) | P00355 | VGVNGMGR-LTGFSMR- QAKNLLPASTGAAK- EPSWYDNEFGYSNR | 43 |
| 37736/51471 | Rubisco large chain (fragment) (Vitis vinifera)f | Q6ZYB1 | LTYYTPEYETKPTDILAAFR-IPPAYSK- TFQGPPHGIQVER-AVYECLR- GGLDFTK-GGLDFTKDDENVNSQPFMR-FLFCAEAIFK-SQAETGEIK- GHYLNATAGTCEEMIK-DNGLLLHIHR | 19 | ||
| GLP6 | Leaf | 38759/43463 | Glycine cleavage complex T-protein (fragment) (Zea mays) | Q8W521 | LTGLGAR-TVLYDFH- LALQGPLAAPVL-DVSWHLHDER | 28 |
| GLP7 | Leaf | 27954/28858 | LHCII type III cab binding protein (Vigna radiata) | gi | 4689380 | WAMLGALGCITPEVLEK- GPLENLLDHLDNPVANNAWVYATK | 15 |
| 27954/10291 | Probable oxygen-evolving enhancer protein 2 (Vitis vinifera)f | gi | 37903240 | TADGDEGGKHQLITAAVADGK- HQLITAAVADGK-KYVESTASSFSIA | 8 | ||
| GLP8 | Leaf | 29850/27033 | Ascorbate peroxidase (Vigna unguiculata) | Q41712 | ELLSGEK-EGLLQLRSDK- GPWTSNPLLFDNSYFK-PEPPPEGR- LLAGPNK-ALLSDPAFRLPVEK | 21 |
| GLP9 | Leaf | 28646/27033 | Ascorbate peroxidase (Vigna unguiculata) | Q41712 | EDKPEPPPEGR- SGFEGPWTSNPLIFDNSYFK ELLSGEK-EGLLQLPSDK | 46 |
Spota . | Tissue . | Massesb . | Identified protein (species) . | Accession no.c . | Matching peptidesd . | %e . |
|---|---|---|---|---|---|---|
| GRP1 | Root | 61804/- | No identification | – | – | – |
| GRP2 | Root | 61804/53394 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (Prunus dulcis) | O24246 | LDQLQLLLK-GVDAQIASGGGR-AVEIAEK-LYEGEGFK | 15 |
| GRP3 | Root | 54911/51887 | UTP-glucose-1P-uridyltransferase (Arabidopsis thaliana) | P57751 | LVEADALK-LVQLETAAGAAIR-SGFINLVSR | 16 |
| GRP4 | Root | 37635/41278 | Orcinol O-methyltransferase (Rosa hybrid) | Q8L5K8 | VIIIDMMMENQK-CTVLDLPHVRAD-FNEAMASDAR-VDVGGGTG- DAGFSGYK-EEGYVLTHAS | 3 |
| GRP5 | Root | 34801/34021 | D-protein (Hordeum vulgare) | Q8VWY8 | SSHALALVGQK-LLSSGDAGPPHR-AYVFGGELTPR | 10 |
| GRP6 | Root | 20651/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 57 |
| GRP7 | Root | 20673/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | GGKEDALATFK-EDALATFK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 58 |
| 20673/37836 | Adenosin kinase 1 (Arabidopsis thaliana)f | Q9SF85 | SLIANLSAAGGCYK-LNNAILAEDK- AGCYASHVPEQR-SFPVLLLPK | 6 | ||
| GRP8 | Root | 18460/- | No identification | – | – | – |
| GRP9 | Root | 19935/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-AAVLDADNLIPK- AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 54 |
| GRP10 | Root | 20710/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | VDAIDKEK-AAVLDADNLIPK- AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 64 |
| GRP11 | Root | 20698/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-VDAIDKEK- GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- CVEVIQGDGGPGTIK- GVFTYEQEITSSVPPAK | 63 |
| GSP1 | Shoot | 37590/- | No identification | – | – | – |
| GSP2 | Shoot | 32385/- | No identification | – | – | – |
| GSP3 | Shoot | 40019/39073 | Glyceraldehyde-3P-dehydrogenase (Atriplex nummularia) | P34783 | VIISAPSK-VLPALNGK-LTGMSFR- SSIFDAK-AASFNIIPSSTGAAK- LVSWYDNEWGYSSR | 25 |
| GSP4 | Shoot | 21618/17419 | Thioredoxin peroxidase 1 (Nicotiana sylvestris) | Q8S310 | HVPGFIEK-VLFGVPGAFT | 11 |
| GSP5 | Shoot | 18929/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | VVASPDGGSIYK-AAILDSDNLIPK- AIEAYVLAHPDAY | 23 |
| GSP6 | Shoot | 18605/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | GGKEDALATFK-VVASPDGGSIYK- AAILDSDNLIPK-GVFTYEQEITSSVPPAK | 32 |
| GSP7 | Shoot | 18345/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | AIEAYVLAHPDAY-AAILDSDNLIPK | 15 |
| GSP8 | Shoot | 19935/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | AIEAYVLAHPDAY-AAVLDADNLIPK- GVFTYEQEITSSVPPAK-GAEVCEEHVK | 15 |
| GSP9 | Shoot | 17211/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | AAVLDADNLIPK | 7 |
| GSP10 | Shoot | 20569/17118 | PRP-10(Vitis vinifera) | Q9FS43 | GGKEDALATFK-GAEVCEEHVK- AAVLDADNLIPK-AIEAYVLAHPDAY- GVFTYEQEITSSVPPAK | 39 |
| GSP11 | Shoot | 20626/17118 | PRP-10 (Vitis vinifera) | Q9FS43 | EDALATFK-AAVLDADNLIPK- AIEAYVLAHPDAY- GVFTYEQEITSSVPPAK | 26 |
| GSP12 | Shoot | 20626/17223 | PRP-10 (Vitis vinifera) | Q9FS42 | AAILDSDNLIPK-AIEAYVLAHPDAY-GVFTYEQEITSSVPPAK | 36 |
| GSP13 | Shoot | 16883/52891 | Rubisco large subunit (fragment) (Lantana camara) | Q9THN3 | DNGLLLHIHR-FLFCAEALFK- GGLDFTK-AETGEIK | 8 |
| GLP1 | Leaf | 54767/51983 | ATP synthase beta subunit (Sparganium americanum) | Q95FJ9 | ESGVINEK-IGLFGGAGVGK-LSIFETGIK- SAPAFIQLDTK-MPNIYNALVVK- TVLIMELINNIAK- IAQIIGPVLDAVFPPGK IFNVLGEPVDNLGPVDTR | 50 |
| GLP2 | Leaf | 32590/34840 | Putative 33 kDa oxygen evolving protein of photosystem II (Oryza sativa) | Q943W1 | FCLEPTSFTVK-VPFLFTIK-GSSFLDPK- GGSTGYDNAVALPAGGR | 20 |
| 32590/34489 | Carbonic anhydrase, chloroplast precursor (Nicotiana tabacum)f | P27141 | YSRGAALEYAVLHLK-YMVFACVCSR | 8 | ||
| GLP3 | Leaf | 40890/40260 | Glycolate oxidase, perisomal (Spinacia oleracea) | Q9FS42 | AIALTVDTPR-IPVFLDGGVR | 5 |
| GLP4 | Leaf | 37334/51471 | Rubisco large chain (fragment) (Bursera inaguensis) | P28385 | IPPAYSK-TFQGPPHGIQVER-AVYECLR- GGLDFTK-FLFCAEALFK-SKAETGELK DNGLLLHLHR | 19 |
| GLP5 | Leaf | 37736/35682 | Glyceraldehyde 3-phosphate dehydrogenase (Sus scrofa) | P00355 | VGVNGMGR-LTGFSMR- QAKNLLPASTGAAK- EPSWYDNEFGYSNR | 43 |
| 37736/51471 | Rubisco large chain (fragment) (Vitis vinifera)f | Q6ZYB1 | LTYYTPEYETKPTDILAAFR-IPPAYSK- TFQGPPHGIQVER-AVYECLR- GGLDFTK-GGLDFTKDDENVNSQPFMR-FLFCAEAIFK-SQAETGEIK- GHYLNATAGTCEEMIK-DNGLLLHIHR | 19 | ||
| GLP6 | Leaf | 38759/43463 | Glycine cleavage complex T-protein (fragment) (Zea mays) | Q8W521 | LTGLGAR-TVLYDFH- LALQGPLAAPVL-DVSWHLHDER | 28 |
| GLP7 | Leaf | 27954/28858 | LHCII type III cab binding protein (Vigna radiata) | gi | 4689380 | WAMLGALGCITPEVLEK- GPLENLLDHLDNPVANNAWVYATK | 15 |
| 27954/10291 | Probable oxygen-evolving enhancer protein 2 (Vitis vinifera)f | gi | 37903240 | TADGDEGGKHQLITAAVADGK- HQLITAAVADGK-KYVESTASSFSIA | 8 | ||
| GLP8 | Leaf | 29850/27033 | Ascorbate peroxidase (Vigna unguiculata) | Q41712 | ELLSGEK-EGLLQLRSDK- GPWTSNPLLFDNSYFK-PEPPPEGR- LLAGPNK-ALLSDPAFRLPVEK | 21 |
| GLP9 | Leaf | 28646/27033 | Ascorbate peroxidase (Vigna unguiculata) | Q41712 | EDKPEPPPEGR- SGFEGPWTSNPLIFDNSYFK ELLSGEK-EGLLQLPSDK | 46 |
Spots are named accordingly to Fig. 1.
Observed molecular mass determined on the gel (Daltons)/Theoretical molecular mass (Daltons).
Accession number in NCBI or SWISS-PROT databases and organism assignment after BLAST homology searches.
Peptides identified via the Mascot search engine and confirmed by de novo sequencing are indicated in regular characters, whereas peptides only identified by de novo sequencing and BLAST search are in bold and italic.
Percentage of amino acids in reference proteins covered by matching peptides from nano-LC-MS/MS analysis.
Co-migrating protein. Abbreviations: GRP, Grapevine Root Protein; GSP, Grapevine Shoot Protein; GLP, Grapevine Leaf Protein.
Further sequencing analysis and subsequent database matching of shoot protein spots resulted in the identification of the enzymes glyceraldehyde-3P-dehydrogenase (GAPDH) and peroxiredoxin 1 (Prx1), which showed coverage scores of 39% and 11%, respectively. Undoubtedly, the most striking result arises from the presence of up to eight proteins that were homologous to two different clones of a PRP-10 from grapevine. However, coverage percentages were significantly lower compared with roots and ranged between 7% and 39%. In addition, spot GSP13 was identified as a fragment of the large subunit of the Rubisco enzyme, although matched peptides only covered 8% of the protein sequence. Spots called GSP1 and GSP2 could not be identified because of the low signal in the MS analysis.
These findings also revealed the putative nature of all proteins induced by fmx in grapevine leaves. The enzymatic fragmentation of spot GLP1 allowed 50% of its amino acid sequence to be matched with an ATP synthase (ATPase) beta subunit. In addition, GLP2 matched to a chloroplast precursor of a carbonic anhydrase (CA) enzyme and a putative 33 kDa oxygen evolving protein of photosystem II (8% and 20% of coverage, respectively), whereas a homologue of the enzyme glycolate oxidase (GO) was also identified from GLP3. Although experimental and theoretical masses were very similar, the coverage score reported (5%) was low. Two spots (GLP4 and GLP5) demonstrated significant coverage scores (19%) with the large chain of the enzyme Rubisco. The spot designated GLP5 covered 43% of the enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Moreover, GLP6 was found to be homologous to a glycine cleavage complex T protein (28% coverage), whereas GLP7 displayed significant coverage percentages with a chlorophyll a/b binding protein and a putative oxygen-evolving enhancer protein 2 (15% and 8% coverage, respectively). Finally, sequence comparisons of spots GLP8 and GLP9 showed 21% and 46% coverage with two ascorbate peroxidases (APX), which play a major role against oxidative stress.
Discussion
Since pesticide stress has not yet been extensively examined at the proteome level, no data on this topic are currently available in the literature. Therefore, this study provides the first basic data and a framework for further investigations. Since field environment fluctuations can induce a number of stresses that may partially mask the herbicide effects, all experiments were performed under optimal growing conditions, using in vitro-grown plantlets. However, although there are obvious advantages of this experimental model, the results cannot immediately be translated to grapevine crop plants, and they will need further verification in the vineyard. The identification strategy was not trivial and required the use of de novo sequencing techniques to identify some of the protein functions by homology in phylogenetically close organisms that are already sequenced. Therefore, this study constitutes a relevant example of the possibility of performing proteomic studies on genomes that have not yet been sequenced. A total of 33 spots showing a differential expression pattern in fmx-stressed tissues compared with controls were studied here. However, a more detailed analysis over the few hundreds of spots firstly scrutinized could enlarge the map of fmx-responsive proteins in future work. Protein fluctuations were first detected in roots 24 h after the onset of treatments, suggesting a rapid plant response to counteract the detrimental effects of fmx. In addition, grapevine proteome expression was differentially altered in roots, shoots, and leaves, suggesting the existence of tissue-specific responses as a consequence of distinct physiological alterations. Here, proteins have been grouped and discussed according to their putative biological functions, assigned on the basis of their sequence homology with known proteins present in databases.
PR-10 proteins
Noteworthy was the accumulation of a number of PRP-10 protein in grapevine roots and shoots in response to fmx stress. Beside their constitutive expression in plants (Wu et al., 2003), PRP-10 are further induced by pathogen infection (Park et al., 2004), wounding (Poupard et al., 2003), drought stress (Dubods and Plomion, 2001), UV exposure (Pinto and Ricardo, 1995), and in the presence of gaseous air pollutants (Pääkkönnen et al., 1998; Agrawal et al., 2002; Rakwal et al., 2003) or heavy metals (Utriainen et al., 1998; Hajduch et al., 2001), suggesting a general defence role for this group of proteins. They are usually members of a multigene family (Wen et al., 1997), so the presence of several spots in roots and shoots argues well with this fact, although the existence of allelic variations should also be considered (Swoboda et al., 1995). Likewise, a recent study has evidence that the PR-10c protein of birch undergoes post-translational modifications (Koistinen et al., 2002), allowing the supposition that some of the spots found here could correspond to processed polypeptides. Further MS analysis will allow this hypothesis to be confirmed. Although the exact catalytic function of most of these proteins in the plant defence response system is as yet unknown, several PRP-10 proteins possess ribonuclease activity in vitro (Bufe et al., 1996; Bantignies et al., 2000; Biesiadka et al., 2002; Zhou et al., 2002; Wu et al., 2003) and in vivo (Park et al., 2004). Further enzymatic assays will be necessary to determine whether any of the PRP-10 proteins detected in fmx-stressed grapevine plantlets possess ribonuclease activity.
Photosynthesis-related proteins
As expected from its mode of action, fmx has recently been found to cause a significant decrease in leaf gas exchange, as well as in the content of both chlorophylls and carotenoids, resulting in a strong inhibition of plant growth and biomass production (Saladin et al., 2003a). In addition, plastids were also structurally altered, showing less developed grana and tylakoid disorganization. Using these results, several spots in shoots and leaves have been identified as the large chain of the Rubisco enzyme. Nevertheless, with regard to their experimental molecular masses, they should be considered as breakdown products. Prominent reduction and/or fragmentation of this major photosynthetic enzyme have also been reported in plants exposed to environmental pollutants (Hajduch et al., 2001; Agrawal et al., 2002; Rakwal et al., 2003) and drought conditions (Costa et al., 1998; Salekdeh et al., 2002a). Although Rubisco fragmentation could also occur in vitro during protein solubilization, the absence of such fragments in control samples makes this hypothesis unlikely here. Other photosynthesis-related proteins affected by the herbicide included two induced leaf proteins, partially identified as a putative oxygen-evolving enhancer protein and an LHCII type III chlorophyll a/b binding protein, respectively. The first protein is of crucial importance for O2 evolution and photosystem (PS) II stability (Sugihara et al., 2000). The second one is a major component of light-harvesting antennae complex of PSII in higher plants, which alleviates excitation energy pressure and avoids photodamage (Anderson et al., 1995). Similarly, the expression of the homologous LHCII gene in the algae Dunaliella tertiolecta was enhanced after exposure to the herbicide dichlorophenyldimethylurea, a photosynthetic electron-transport inhibitor (Escoubas et al., 1995). Therefore, these data here suggest that fmx could interfere negatively with photosynthetic activity in grapevine plantlets.
Enzymes involved in photorespiration
In a recent work (Saladin et al., 2003a), flumioxazin was shown to cause a decrease in the water content and water potential of shoots and leaves. It is well known that water losses in plants lead to the closure of stomata, causing a diminution in CO2 levels. A low CO2 environment could activate photorespiration (Noctor et al., 2002), which is widely accepted to function as an alternative electron sink, making photosynthesis less efficient. This effect is of great advantage under unfavourable conditions (e.g. water shortage), diminishing the possibility of photoinhibition and mitigating the formation of active oxygen species (AOS) in chloroplasts (Osmond and Grace, 1995). Two key enzymes involved in photorespiration, GO and glycine cleavage T protein, were partially identified in fmx-stressed leaves. The increase detected for both proteins suggests the stimulation of this metabolic pathway in fmx-stressed grapevine leaves. In addition, another leaf protein was partially homologous to a chloroplast precursor of a carboxylase enzyme. Within the C3 chloroplast, this enzyme could play a direct role in the supply of CO2 for Rubisco when its availability is limited (Raven, 1997).
Antioxidant enzymes
Damage and/or disturbances of the photosynthesis pathway could lead to AOS production, causing detrimental effects to cellular functions (Foyer et al., 1994). In a recent study, transient accumulation of thiobarbituric acid reactive substances (TBARS) and significant variations of the relative electrolyte leakage in fmx-treated grapevine leaves were reported (Saladin et al., 2003b). These findings suggest that lipid peroxidation could occur in foliar cell membranes, probably as a result of an increase in AOS levels. Antioxidant enzymes are relevant endpoints among the defence mechanisms against oxidative stress. Accordingly, two induced proteins from stressed grapevine leaves exhibited a significant coverage with two APXs. These enzymes link the detoxification of toxic H2O2 to oxidation of the reductant ascorbate (Asada, 1992). The presence of two different protein spots argues well with the several APX isoforms previously reported (Asada, 1992). Another induced protein in the shoots was partially identified as a peroxiredoxin enzyme (Prx1). These ubiquitous enzymes catalyse the destruction of H2O2 and alkyl hydroperoxides using thioredoxin as the electron donor. Experimental evidence suggests that peroxiredoxins may have a protective role in the context of photosynthesis (Dietz, 2003).
Enzymes of sugar metabolism
It is now well known that plant resistance to stress factors is tightly connected with energy metabolism. Indeed, variations in carbohydrate concentration have often been implicated in the responses to a variety of stresses (Roitch, 1999). Several proteins involved in sugar metabolism were partially identified in fmx-treated grapevine tissues, varying their expression patterns after herbicide exposure. Thus, a putative homologous protein of the enzyme PGAM-i was significantly repressed in fmx-exposed grapevine roots. This could lead to an increased pool of 3-phosphoglycerate, reversing the glucose catalytic flux towards the biosynthetic pathway Another protein spot showing significant homology with a UGPase enzyme was induced in stressed roots. The increasing levels of both enzymes under herbicide stress could contribute to explain the rise in soluble sugar contents found by (Saladin et al., 2003a, b). In addition, a homologous protein of the enzyme GAPDH was newly synthesized in fmx-treated shoots and leaves. This enzyme is part of the so-called anaerobic proteins, which are selectively synthesized under O2 deprivation (Sachs et al., 1980) allowing plant survival when oxygen is limited. Stomata closure due to water deficit, as well as a decrease in O2 production owing to an impaired photosynthesis (Saladin et al., 2003b), could induce GADPH synthesis in fmx-stresssed shoots and leaves. Similarly, the enzyme has been detected to accumulate in leaves of Mesembryanthemum crystallinum exposed either to saline or drought conditions (Forsthoefel et al., 1995). However, anaerobic proteins, including GAPDH, do not seem to be key regulatory enzymes in glycolysis (Miernyk, 1990). Therefore, more protein does not necessarily mean an enhanced metabolic flux. In this sense, any other quite different biological role for this enzyme under fmx stress cannot be dismissed. Because of the major function of all these enzymes in sugar metabolism, their variations due to the herbicide treatment presumably reflects altered patterns of carbon flux in response to reduced photosynthesis and an increased need for osmotic adjustment in grapevine tissues.
Concluding remarks
Several conclusions can be drawn in the light of the findings reported here. First of all, these results are in good agreement with previous work conducted at a physiological level and allow protein changes to be associated with physiological traits affected by fmx. Moreover, the existence of significant changes in the proteome of all the organs analysed, together with previous physiological data (Saladin et al., 2003a, b), suggests that the herbicide could act systemically in grapevine tissues, probably via root uptake. Finally, some of these proteins could serve as putative biochemical markers to monitor the presence of the herbicide in grapevine tissues, as well as to evaluate the impact of fmx in some other non-target species of the vineyard ecosystem.
Future work will be aimed at: (i) examining the effects of fmx on soil-grown plants at proteome level, (ii) determining whether the observed protein variations reflect changes in gene expression, and (iii) determining the biological role of grapevine PRP-10 proteins in the defence response against fmx.
Answering these and other questions will improve the understanding of fmx responsiveness in grapevine and could eventually lead to applications in breeding for enhanced pesticide tolerance. Moreover, since this work brings some evidence of the detrimental effects of fmx on grapevine physiology, it could contribute to a more environment friendly use of these chemicals in future.
Present address: Departamento de Bioquímica, Biología Celular y Molecular de Plantas, Estación Experimental del Zaidín (CSIC), C/ Profesor Albareda 1, E-18008 Granada, Spain.
Present address: LEBHAM, UPRES EA 3877, Institut Universitaire Européen de la Mer, Université de Bretagne Occidentale, Technopole Brest Iroise, Place Nicolas Copernic, F-29280 Plouzané, France.
Abbreviations: AOS, active oxygen species; APX, ascorbate proxidase; CA, carbonic anhydrase; fmx, flumioxazin; GAPDH, glyceraldehyde-3P-dehydrogenase; GLP, grapevine leaf protein; GO, glycolate oxidase; GRP, grapevine root protein; GSP, grapevine shoot protein; LHC, light-harvesting complex; PGAM-i, 2,3 biphosphoglycerate-independent phosphoglycerate mutase; PR, pathogenesis-related; Prx, peroxiredoxin; PS, photosystem; Rubisco, ribulose-1,5-biphosphate carboxylase/oxygenase; UGPase, UTP-glucose pyrophosphorylase.
This project was partially funded by Europol'Agro (Reims, France). We are also grateful to the Bruker Daltonics Company and the CNRS (France) for supporting C Carapito with a fellowship.
References
Agrawal GK, Rakwal R, Yonekura M, Kubo A, Saji H.
Anderson JM, Chow WS, Park YI.
Anderson N, Esquer-Blasco R, Hoffmann JP, Anderson NG.
Asada K.
Bantignies B, Seguin J, Muzac I, Dedaldechamp F, Gulick P, Ibrahim R.
Biesiadka J, Bujacz G, Siorski MM, Jaskolski M.
Bradford M.
Bufe A, Spangfort MD, Kahlert H, Schlaak M, Becker WM.
Chang WWP, Huang L, Shen M, Webster C, Burlingame AL, Roberts JKM.
Costa P, Bahrman N, Frigerio JM, Kremer A, Plomion C.
Dubods C, Plomion C.
Escoubas JM, Lomas M, LaRoche J, Falkowski PG.
Fecht-Christoffers MM, Braun HP, Lemaitre-Guillier C, VanDorsselaer A, Horst WJ.
Forsthoefel NR, Vernon DM, Cushman JC.
Foyer CH, Lelandais M, Kunert KJ.
Görg A, Postel W, Günther S.
Hajduch M, Rakwal R, Agrawal GK, Yonekura M, Pretova A.
Jame YW, Cesnna AJ, Biederbeck VO, Grover R, Smith AE, Korven HC.
Koistinen KM, Kokko HI, Hassinen VH, Tervahauta AI, Auriola S, Kärenlampi SO.
Laemmli UK.
Lee HJ, Duke SO.
Lund AA, Blum PH, Bhattramakki D, Elthon TE.
Lyndon J, Darlington L.
Martin C, Vernoy R, Carré M, Vesselle G, Collas A, Bougerey C.
Miernyk JA.
Noctor G, Veljovic-Jovanovic S, Driscoll S, Novitskaya L, Foyer CH.
Osmond CB, Grace SC.
Pääkkönnen E, Seppänen S, Holopainen T, Kokko H, Kärenlampi S, Kärenlampi L, Kangasjärvi J.
Park CJ, Kim KJ, Shin R, Park JM, Shin YC, Paek KH.
Pinto MP, Ricardo CPP.
Poupard P, Parisi L, Campion C, Ziadi S, Simoneau P.
Rabilloud T, Brodard V, Peltre G, Righetti PG, Ettori C.
Rakwal R, Agrawal GK, Kubo A, Yonekura M, Tamogami S, Saji H, Iwahashi H.
Rakwal R, Agrawal GK, Yonekura M.
Raven JA.
Riccardi F, Gazeau P, de Vienne D, Zivy M.
Roitch T.
Saladin G, Magné C, Clément C.
Saladin G, Clément C, Magné C.
Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J.
Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J.
Sugihara K, Hanagata N, Dubinsky Z, Baba S, Karube I.
Suzuki K, Itai R, Suzuki K, Nakanishi H, Nishizawa NK, Yoshimura E, Mori S.
Swoboda I, Scheiner O, Heberle-Bors E, Vicente O.
Tomlin CDS.
Ukaji N, Kuwabara C, Takezawa D, Arakawa K, Yoshida S, Fujikawa S.
Utriainen M, Kokko H, Auriola S, Sarrazin O, Kärenlampi S.
Wen J, Vanek-Krebitz M, Hoffmann-Sommerbruger K, Scheiner O, Breiteneder H.
Wu F, Yan M, Li Y, Chang S, Song X, Zhou Z, Gong W.



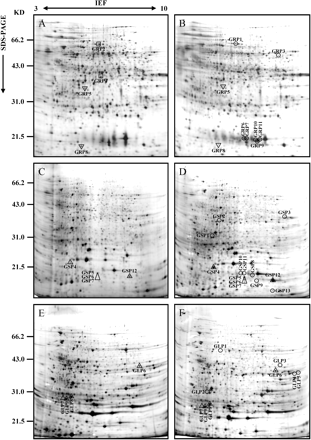
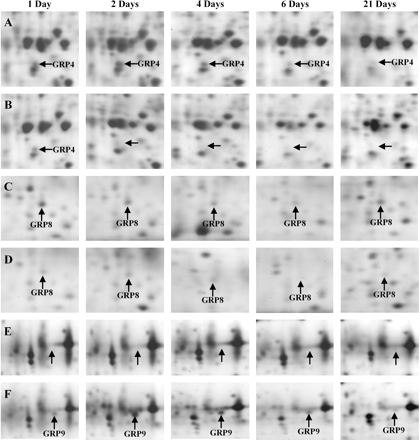
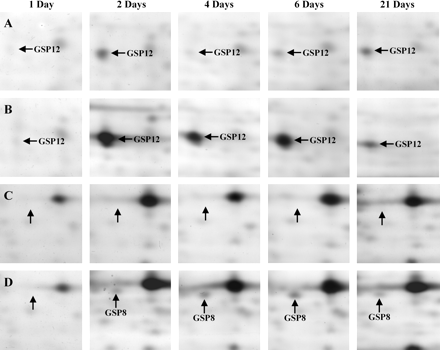
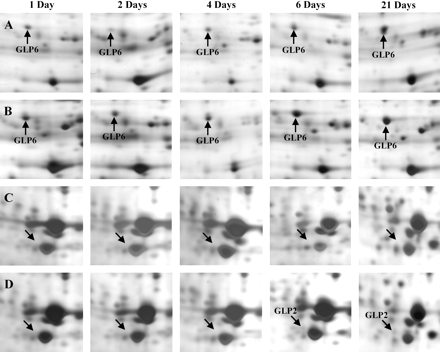

Comments